Citation: Colautti RI, Franks SJ, Hufbauer RA, Kotanen PM, Torchin M, Byers JE, Pyšek P, Bossdorf O (2014) The Global Garlic Mustard Field Survey (GGMFS): challenges and opportunities of a unique, large-scale collaboration for invasion biology. In: Capdevila-Argüelles L, Zilletti B (Eds) Proceedings of 7th NEOBIOTA conference, Pontevedra, Spain. NeoBiota 21: 29–47. doi: 10.3897/neobiota.21.5242
To understand what makes some species successful invaders, it is critical to quantify performance differences between native and introduced regions, and among populations occupying a broad range of environmental conditions within each region. However, these data are not available even for the world’s most notorious invasive species. Here we introduce the Global Garlic Mustard Field Survey, a coordinated distributed field survey to collect performance data and germplasm from a single invasive species: garlic mustard (Alliaria petiolata) across its entire distribution using minimal resources. We chose this species for its ecological impacts, prominence in ecological studies of invasion success, simple life history, and several genetic and life history attributes that make it amenable to experimental study. We developed a standardised field survey protocol to estimate population size (area) and density, age structure, plant size and fecundity, as well as damage by herbivores and pathogens in each population, and to collect representative seed samples. Across four years and with contributions from 164 academic and non-academic participants from 16 countries in North America and Europe thus far, we have collected 45, 788 measurements and counts of 137, 811 plants from 383 populations and seeds from over 5, 000 plants. All field data and seed resources will be curated for release to the scientific community. Our goal is to establish A. petiolata as a model species for plant invasion biology and to encourage large collaborative studies of other invasive species.
Alliaria petiolata, citizen science, model system, plant invasion, research network
How is it that invasive species, which are introduced to novel geographical regions where they lack an adaptive evolutionary history, are nonetheless able to establish and proliferate? Since investigation into this topic was launched over a half-century ago (
A key distinction between biogeographical and trait-based hypotheses is whether introduced populations have increased in performance relative to native populations, where performance may be measured as abundance, range size, demography, and individual survival, reproduction or competitive ability. Biogeographical hypotheses that seek to explain how introduced species escape the regulatory mechanisms present in their native range make an inherent assumption that introduced populations benefit from an ecological or evolutionary increase in performance relative to native populations, on average. We call this the “increased vigour assumption”. In contrast, trait-based hypotheses assume that native and introduced populations perform similarly, with invasion success owing to preadaptation and environmental similarities between ranges. Therefore it is possible two very different types of invaders may exist: (i) species that become abundant and widespread through niche expansion (e.g. escaping regulation or gaining access to new resources) and (ii) species that expand their range through human agency but ultimately perform the same in native and introduced regions. Moreover, the species that both perform well in their native range and expand their niche in invaded regions should be the most successful invaders. However, few studies have measured performance of natural populations to directly test the assumption of increased vigour and these have been limited in size and geographical scope (
In addition to testing for increased vigour, direct field measurements help to assess the ecological relevance of factors proposed to explain invasion success. By “ecological relevance” we mean the extent to which factors affecting fitness components measured under experimental conditions directly translate to invasion success in natural populations and at larger biogeographical scales. Field measurements of natural populations are important because complex interactions among ecological and evolutionary processes can limit the predictive power of ecological and genetic factors identified in controlled experiments. For example, many plants and animals have lost specialist enemies following introduction to geographically distant locales (
Testing the assumption of increased vigour, distinguishing biogeographical and phylogenetic effects, and measuring the ecological relevance of hypotheses of invasion success are difficult tasks. Ideally, a study of increased vigour in a single species would involve (i) extensive field surveys measuring performance of native and introduced populations, (ii) large-scale field manipulations to study population dynamics at nested spatial scales, (iii) development of genetic resources, and (iv) an integrated experimental approach involving experts in a variety of areas including chemical ecology, community dynamics, population ecology, population genetics, developmental biology, and genomics. To facilitate such an approach, our goal is to build a model species for invasion biology, develop novel resources and encourage international and interdisciplinary collaboration to coordinate detailed and robust research on a single species: garlic mustard, Alliaria petiolata. Such an approach could then be applied to additional species in the future. Below we review our motivation for the project, introduce the sampling protocol, describe constraints on protocol design and implementation, summarize the extent of participation, outline our curation and quality control procedures, and note potential avenues for future research.
The Global Garlic Mustard Field Survey (GGMFS) was conceived during a 2008 meeting of the Global Invasions Network, funded by a Research Collaboration Network grant from the U.S. National Science Foundation (see Acknowledgements). The meeting involved 35 invasion biologists representing a broad range of empirical and theoretical backgrounds in ecology, evolution and genetics. Discussions within this group identified the need for standardized field measurements of performance traits, and led to our choice of study species and experimental design.
A major challenge in ecology is generalizing from individual studies done under controlled conditions or in particular locations to broad-scale ecological patterns. This is particularly difficult in invasion biology where ranges often span continents and resources are split among studies of hundreds of different invasive species. Just as large genomic databases and bioinformatics techniques are advancing scientific understanding of genetics and evolution, large-scale ecological data collection networks that focus on a common goal can provide comprehensive data to improve understanding of ecological processes and interactions (
Pure citizen-science projects like project bud-burst (http://budburst.org) and the Evolution MegaLab (http://www.evolutionmegalab.org) (
Large-scale collaborative research projects involving hundreds of scientists create a number of specific challenges, trade-offs and financial considerations. Development of a standardized protocol for large distributed studies requires decisions about the intensity of study and sophistication of participants (Figure 2). Given limited human resources and time, increased intensity of sampling at each site will trade off with the number of sample sites. Additionally, more sophisticated measurements require more expertise, reducing the number of qualified participants. In contrast to most citizen science projects, which usually include very simple measurements (e.g. presence of a species) with little or no equipment or training, we developed a protocol that requires only basic measuring supplies, but can be slightly challenging for participants with no formal science education, and can take several hours to sample a dense population. Nonetheless, we deliberately excluded more complicated but time-intensive field manipulations or measurements such as survival rates, seed production, edaphic measurements, and community composition to encourage an increased number of participants and to keep the protocol accessible to non-scientists. As a result, GGMFS participants are mostly professionally-trained, full-time scientists at academic and governmental organizations who are aware of the need for careful and unbiased data collection. A smaller number of populations have been sampled by non-scientists, and these will be analysed for potential data quality issues in future analyses.
Garlic mustard, Alliaria petiolata (M. Bieb.) Cavara & Grande, is native to most of Europe and western Asia below 68°N latitude (
Herbarium records (
Invasive populations of Alliaria petiolata have been studied frequently. In North America, the species lacks its native specialist herbivores (
Alliaria petiolata is a biennial monocarpic species. Seeds germinate in spring, overwinter as rosettes, flower the following spring, and produce fruits in early summer (
Alliaria petiolata is the sole member of its genus found in Europe and North America (
Diagnostic characteristics of Alliaria petiolata. A populations B rosettes C bolting inflorescences and D individual flowers and developing siliques.
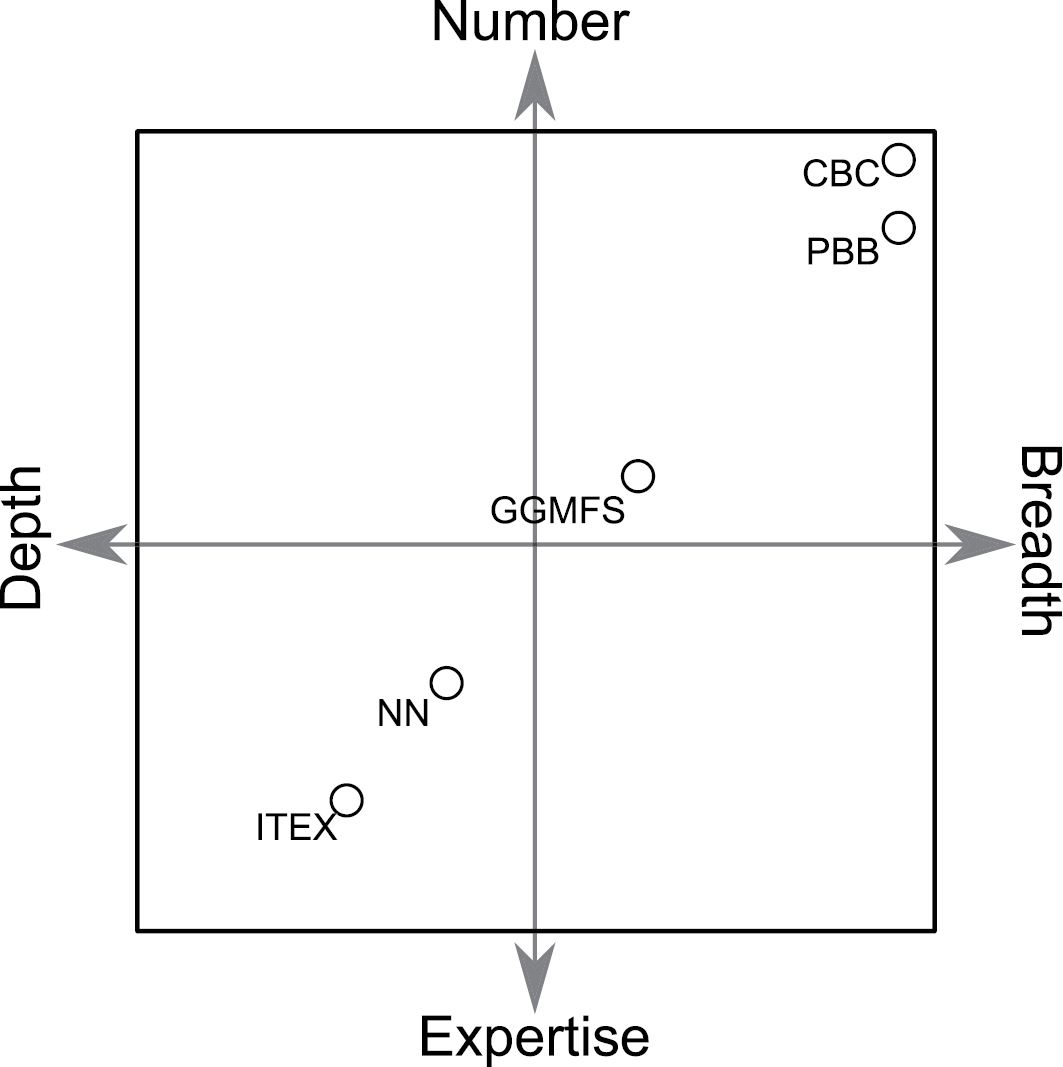
Schematic showing two hypothetical trade-off axes: 1. (x-axis) Sampling intensity: depth of data collection (e.g. number of measurements per site) increases towards the left while the breadth of coverage (e.g. number of sites) increases towards the right; 2. (y-axis) Participant sophistication: number of participants increases towards the top while the average level of expertise per participant increases toward the bottom. Approximate position of the Global Garlic Mustard Field Survey (GGMFS) is shown in relation to other large collaborations in ecology: Nutrient Network (NN); National Ecological Observatory Network (NEON); International Tundra Experiment (ITEX); and the Alpine Stress Gradient Project (ASG), and to citizen science projects: Christmas Bird Count (CBC) and Project Budburst (PBB).
The Brassicaceae is a genetically well-studied family of angiosperm. It includes many economically important horticultural and crops species and the model species Arabidopsis thaliana, which diverged from a common ancestor with Alliaria petiolata only about 20 million years ago (
We established the GGMFS in 2009 to measure population size (i.e. area of coverage), density and age structure (i.e. proportion of first-year rosettes vs. second-year reproductive adults), as well as size, reproduction, herbivory and fungal damage of individual plants, plus some important environmental variables such as habitat type and canopy cover. The current full protocol and data collection sheet are available at http://www.garlicmustard.org, archived on FigShare (https://doi.org/10.6084/m9.figshare.729274), and are described briefly below.
Contributors to the GGMFS are expected to sample populations in the late spring or early summer after >95% of plants have finished flowering. The specific date varies by climate because sampling is intended to standardize phenology across populations and to collect mature seeds for future experiments. We generally encourage participants to include one site with low light (e.g. a forest interior location), and one with high light (e.g. forest edge or roadside). However, to limit selection bias, any site containing 20 or more Alliaria petiolata plants can be included in the study.
Measurements begin by pacing out the approximate area of the population of A. petiolata (length × width). The participants then lay out a 10 m transect to measure plants from one edge of the population moving toward the centre. In each of 10 adjacent 1 × 0.5 m plots, all juvenile rosettes and adult plants are counted. The size of the nearest juvenile and adult plant, as well as the height, leaf number and fecundity (number of fruits) of the adult are measured at five intervals, 20 cm apart, along each plot. On the adult plants, participants also count both the number of undamaged leaves and the number of leaves with >10% herbivore damage. In 2011, we added a simple measurement for pathogen damage, by noting both the total number of plants in each plot that exhibited signs of leaf pathogens as well as the number of damaged leaves. To estimate % canopy cover at each site, photos are taken of the forest canopy at three points across the sampled transect and a visual estimate of average cover is recorded. We use digital camera photos for these estimates because they are common equipment whereas a fish-eye lens is a more accurate but specialized piece of equipment that many people may not have access to. The participants also note whether there is any information of past or ongoing disturbance or control efforts applied to the population they are sampling.
After measurements are recorded, participants harvest inflorescences from the first 20 adult plants in each population. These collections are dried for at least two weeks and then mailed along with the original data sheets and canopy photos. Seed collections and photos of sampled populations provide confirmation that the correct species was sampled, which is of concern mainly for private citizen contributions. Upon receiving these collections we visually inspect the online and hand-written data for potential typographical errors, and we clean and store all seed collections at 5°C. We use ImageJ (
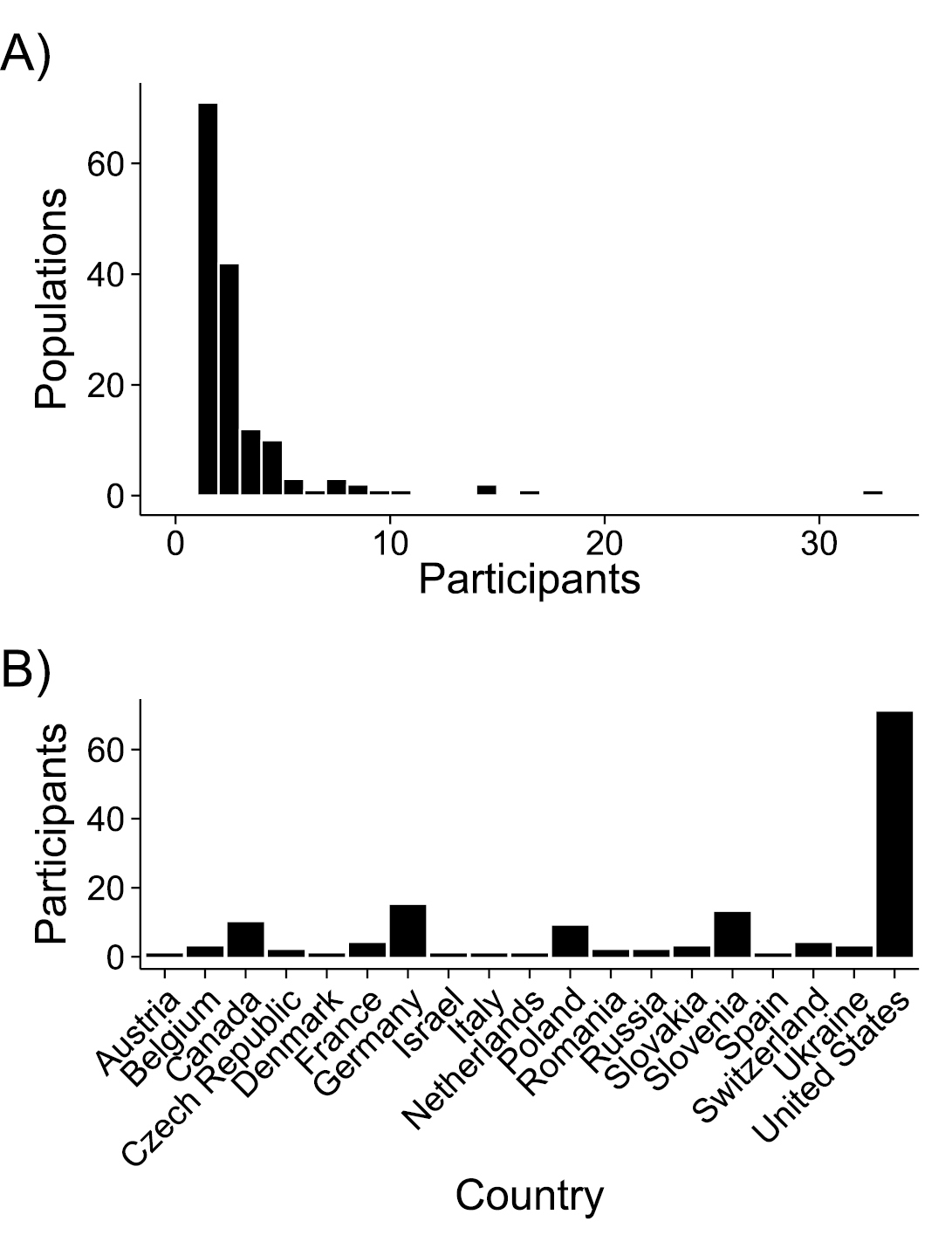
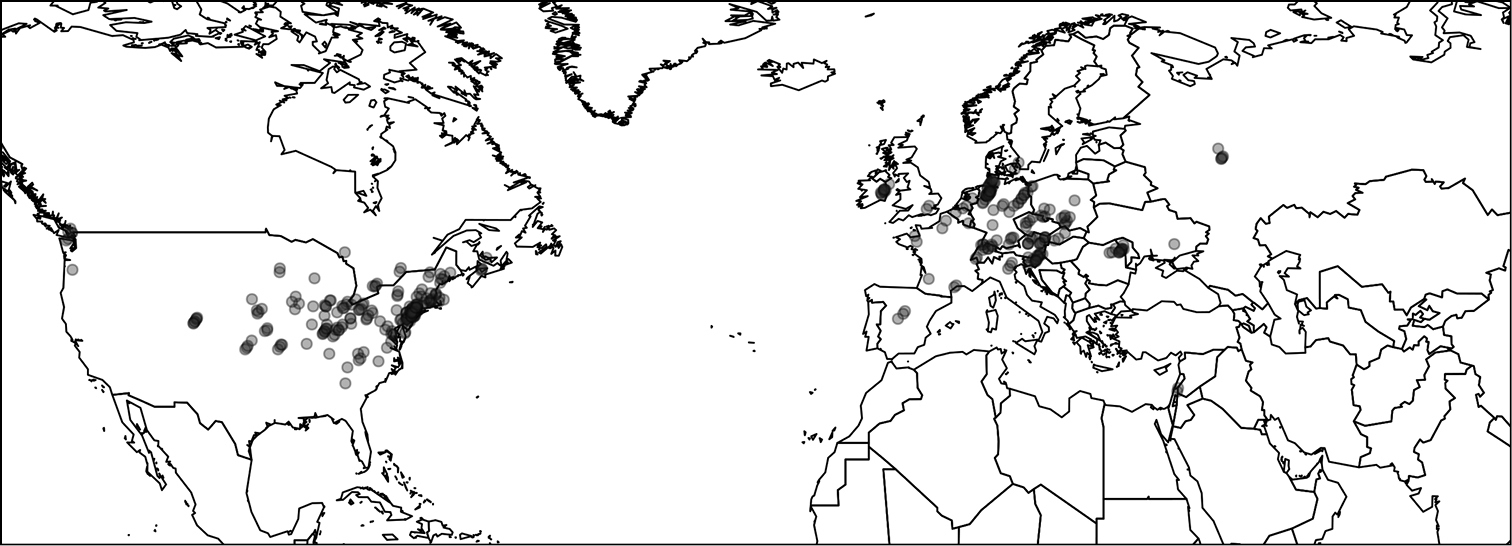
Across four field seasons (2009–2012), 164 participants in 16 countries across North America and Europe (Figure 3) collected data and sampled seeds from 383 field sites (Figure 4). These participants included many academic scientists – faculty, postdocs, and graduate students – but also weed managers, conservation groups, and citizen scientists. Sampling intensity was strongly skewed as most contributors sampled a small number of populations, but a few individuals sampled many sites (Figure 3A). Interestingly, participation at each site was generally all-or-nothing; only five entries were excluded due to incomplete data. Sampling began in 2009 but was most heavily promoted in 2010 and 2011 through direct invitation of individual scientists, as well as general announcements on listserv (e.g. ECOLOG-L, EvolDir), and citizen science channels. After the 2011 season, participants were no longer directly solicited, and participation dropped to 19 populations in 2012. The total field data contributed thus far includes 383 populations concentrated in the northeast U.S. and central Europe (Figure 4). These samples represent a fair proportion of North American and European distribution of Alliaria petiolata (
Frequency histograms of A sampling effort by each participant, and B number of participants per country. Note that “participant” in this case is either an individual or a group of people sampling together.
Map showing 383 sample locations from 2009–2012 inclusive, representing both the native (Europe) and introduced (North America) ranges. Dots are translucent resulting in darker areas that indicate regions with higher sampling intensities.
The GGMFS protocol has been incorporated into some undergraduate-level field courses, particularly at universities focused on undergraduate teaching and educating minorities. These faculty positions are typically associated with high teaching loads and low research budgets, which limit research output relative to faculty at major universities. At the same time, these schools tend to have smaller class sizes and more involved undergraduate students. Large collaborative networks like the GGMFS allow faculty with limited time and financial resources to collaborate within a global community of researchers and play a key role in collecting and analysing valuable scientific data (
In addition to involving faculty in research, the GGMFS data and protocol introduce students to research and provide excellent opportunities for project-oriented undergraduate teaching. Active learning exercises, in which students engage directly in laboratory or field demonstrations, are more effective than class lectures and other passive modes of teaching (
One of the guiding principles of the GGMFS is the inherent value to society of completely open and accessible data, analyses, germplasm and any additional resources arising from this project. Creating these resources is also a crucial step for cultivating a diverse community of biologists with a variety of expertise but a common goal of understanding the biological mechanisms underlying the invasion success of an ecologically important invasive species. We plan to eventually release all data collected for the GGMFS, along with relevant analyses needed to replicate any results published in peer reviewed journals. These will be available to the scientific community with the expectation that future analyses using the data will also be made equally open-access. The release of the field data will likely occur in a series of stages following publication, with a sequester period of 1–2 years to facilitate novel analyses among project participants before releasing data to the scientific community.
Seeds collected as part of the field survey will form the basis of inbred lines for laboratory demonstrations and for future research. Seed collections are stored under cool-dry conditions in three locations – University of Tübingen, Germany, Fordham University in New York, USA, and the University of British Columbia in Vancouver, Canada. To reduce the potential for maternal effects and to produce enough seeds for future experiments, we are currently propagating all viable seed families in an outdoor common garden in Tübingen.
In addition to collecting valuable data and seed resources, the GGMFS has brought together a global network of scientists who posess a range of expertise and a unifying interest in understanding biological invasions. To build on the work described above, we propose to expand this network and to consider additional studies that complement the GGMFS data and address difficult but important questions about the ecology and evolution of invasive species. Anyone interested in participating should contact the lead authors of this paper or the project coordinators listed on the GGMFS website (http://www.garlicmustard.org). We have identified three projects in particular that build on the strengths of the GGMFS model:
(i) Temporal sampling: More detailed demographic measurements, and long-term sampling of the same populations across multiple years would help improve understanding of invasion dynamics at several nested spatial scales.
(ii) Additional invasive species: Replicating the GGMFS approach with other invasive species, including those with different life-history strategies and extent of invasiveness, would allow for a more general test of the increased vigour assumption and testing hypotheses of invasion success in other species. In addition to all of the benefits described above for Alliaria petiolata, field surveys from multiple species will help to identify generalities in the relative importance of different ecological and evolutionary processes to invasion success.
(iii) Large-scale reciprocal transplant experiments: Reciprocal transplant experiments have a long history in plant biology but have rarely been used to study invasive species. Utilizing the GGMFS network and additional collaborators, a large transcontinental reciprocal transplant experiment across dozens of sites in North America and Europe and using a shared subset of GGFMS seed families would be particularly useful to test for genetic differences between, and local adaptation of, native and introduced populations.
Large collaborations are transforming many areas of science, but ecological and evolutionary studies of invasive species have spread limited resources across a broad range of study systems and geographic locations. Focusing studies on a few model systems can help to comprehensively address the fundamental question of what determines invader success, and to evaluate different mechanisms of invasiveness. The Global Garlic Mustard Field Survey (GGMFS) is a step toward this more integrated approach to invasion biology; it provides much-needed comprehensive data on the performance of natural populations of an invasive species across its native and introduced ranges. Large field surveys can provide an important link from experimental results observed at local sites on a subset of populations to biogeographical patterns of invasion success occurring at continental and global scales.
This project was initiated during a Global Invasions Network (NSF RCN) meeting supported by NSF DEB Grant No. 0541673 and benefited from discussions with S. Atkinson, H. Auge, A. Blair, D. Blumenthal, C. Brown, H. Cornell, R. Holt, V. Jarošík, A. Kanarek, J. Kolbe, M. Lampo, C. Lee, L. Martin, W. Miller, C. Mitchell, H. Müller-Schärer, W. Nentwig, V. Panov, J. Parker, J. Pergl, G. Ruiz, U. Schaffner, K. Schierenbeck, R. van Klinken, K. Vaughn, M. Vilà, C. Webb and L. Wolfe. RIC is grateful for fellowship funding from Canadian NSERC-PGS and NSERC-Banting programs, and PMK is supported by an NSERC Discovery Grant. RAH acknowledges support from the Colorado State University Agricultural Experiment Station. PP was supported by long-term research development project no. RVO 67985939 from the Academy of Sciences of the Czech Republic, institutional resources of Ministry of Education, Youth and Sports of the Czech Republic, and acknowledges the support by Praemium Academiae award from the Academy of Sciences of the Czech Republic.