|
||
|
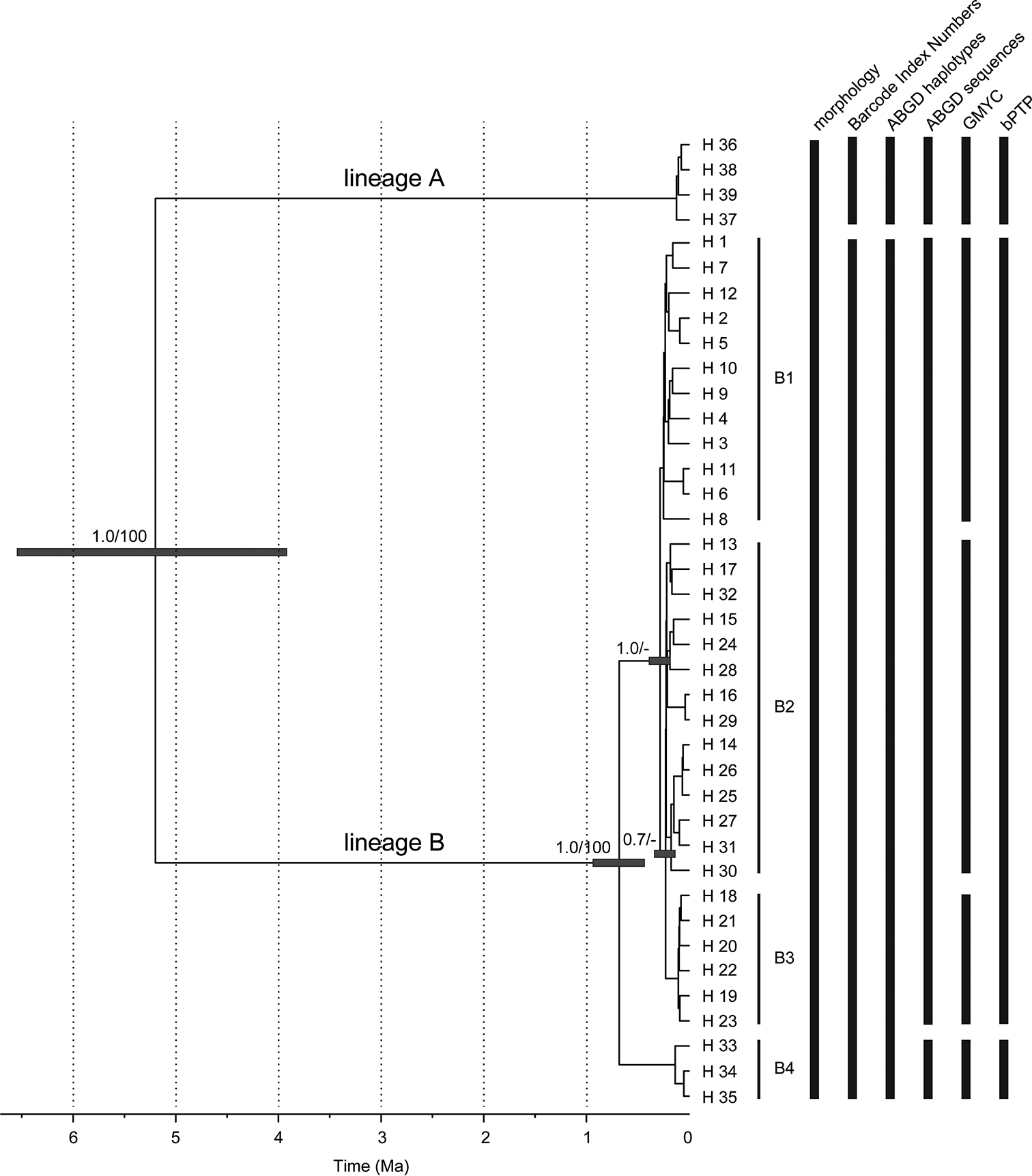
Time-calibrated phylogeny of Dikerogammarus haemobaphes and the results of different species delimitation methods. Maximum clade credibility chronogram was inferred from a strict molecular clock model based on the COI + 16S data set of D. haemobaphes. The numbers given next to the respective main nodes indicate Bayesian posterior probabilities (> 0.5) and ML bootstrap values (> 50%). B1-4 represent the geo-demographic units within the lineage B. BIN – Barcode Index Number (BIN) System, ABGD haplotypes – Automatic Barcode Gap Discovery based on haplotypes, ABGD sequences – Automatic Barcode Gap Discovery based on all sequences, GMYC – Generalized Mixed Yule Coalescent, bPTP – Bayesian Poisson Tree Processes. |