|
||
|
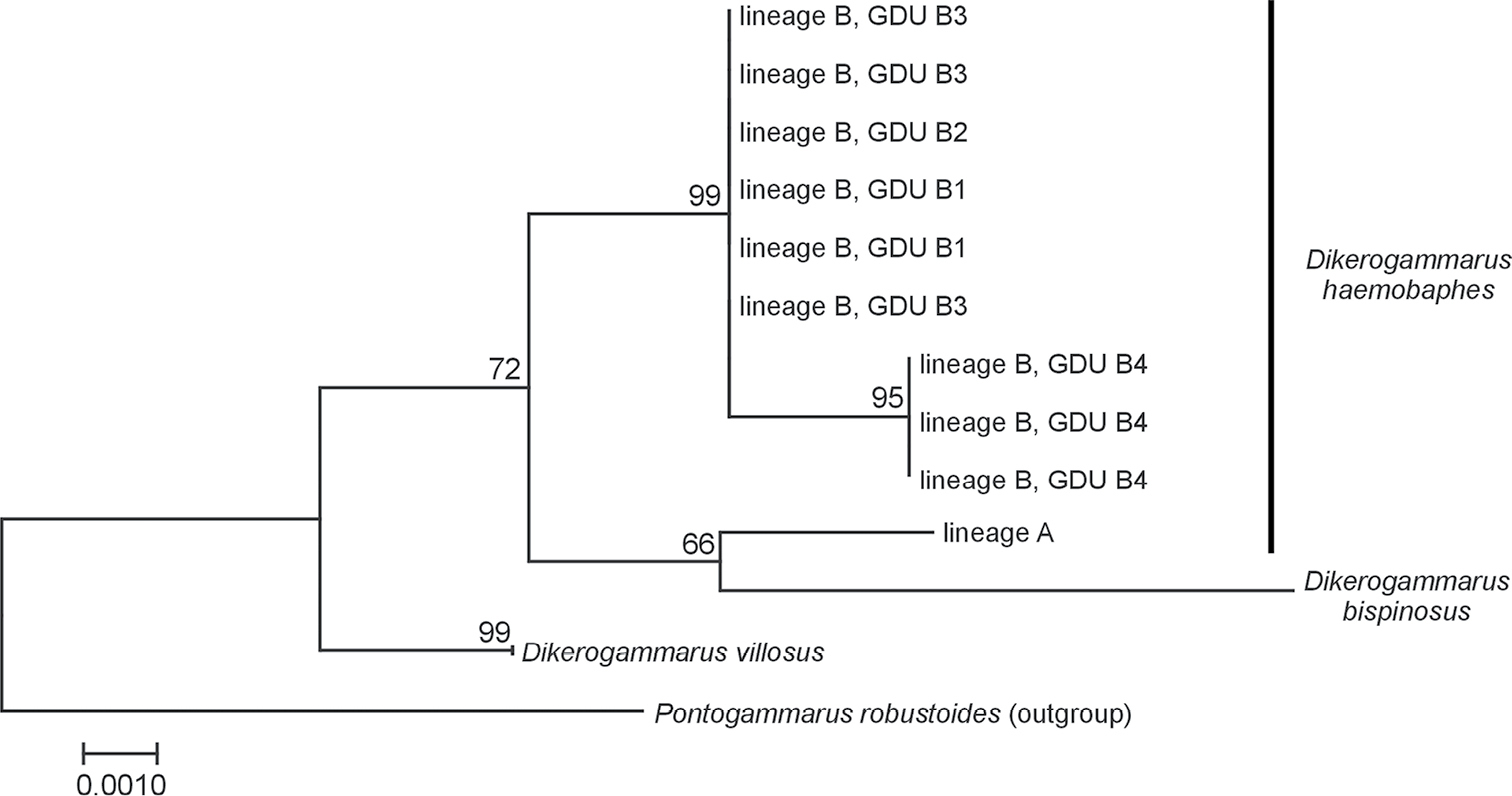
Maximum Likelihood tree of Dikerogammarus spp. based on the 28S rRNA gene. Sequences representing D. haemobaphes lineage A, each of the geo-demographic units (GDU B1-4) within D. haemobaphes lineage B as well as D. bispinosus and D. villosus were used. The evolutionary history was inferred by using the Maximum Likelihood method and the Tamura 3-parameter model. All positions with less than 95% site coverage were eliminated. Bootstrap test was applied with 500 replicates. Information about the D. bispinosus sequence may be found in public BOLD dataset: DHAEMOBA. Dikerogammarus villosus and Pontogammarus robustoides sequences were retrieved from GenBank. |