|
||
|
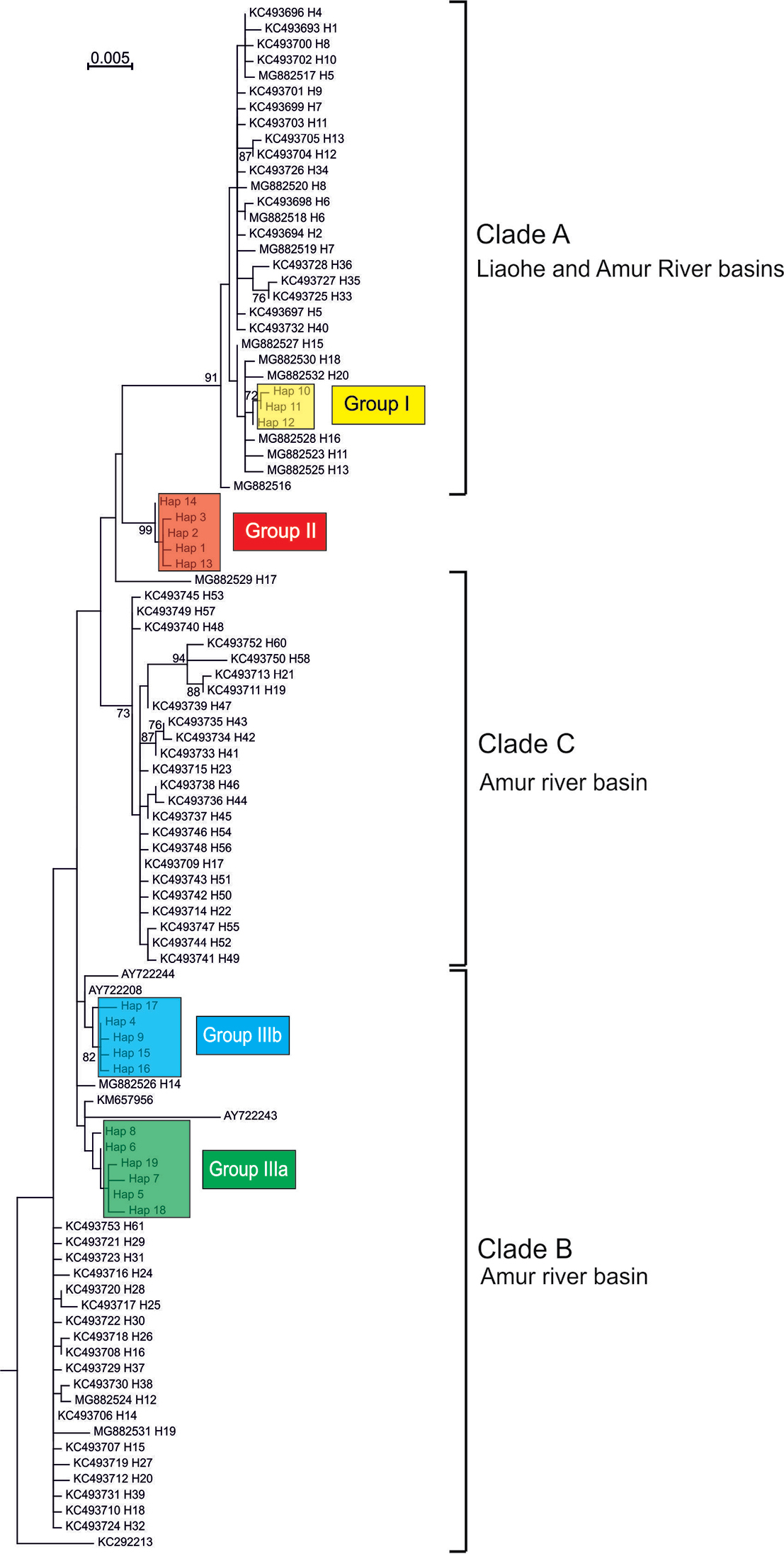
Maximum Likelihood tree showing the relationships of cytochrome b haplotypes identified in our study with the haplotypes from the native area in China (Xu et al. 2013, 2014). Numbers at each node are bootstrap probabilities based on 1000 replications; shown only when they are 70% or higher. Outgroup not shown. |