|
||
|
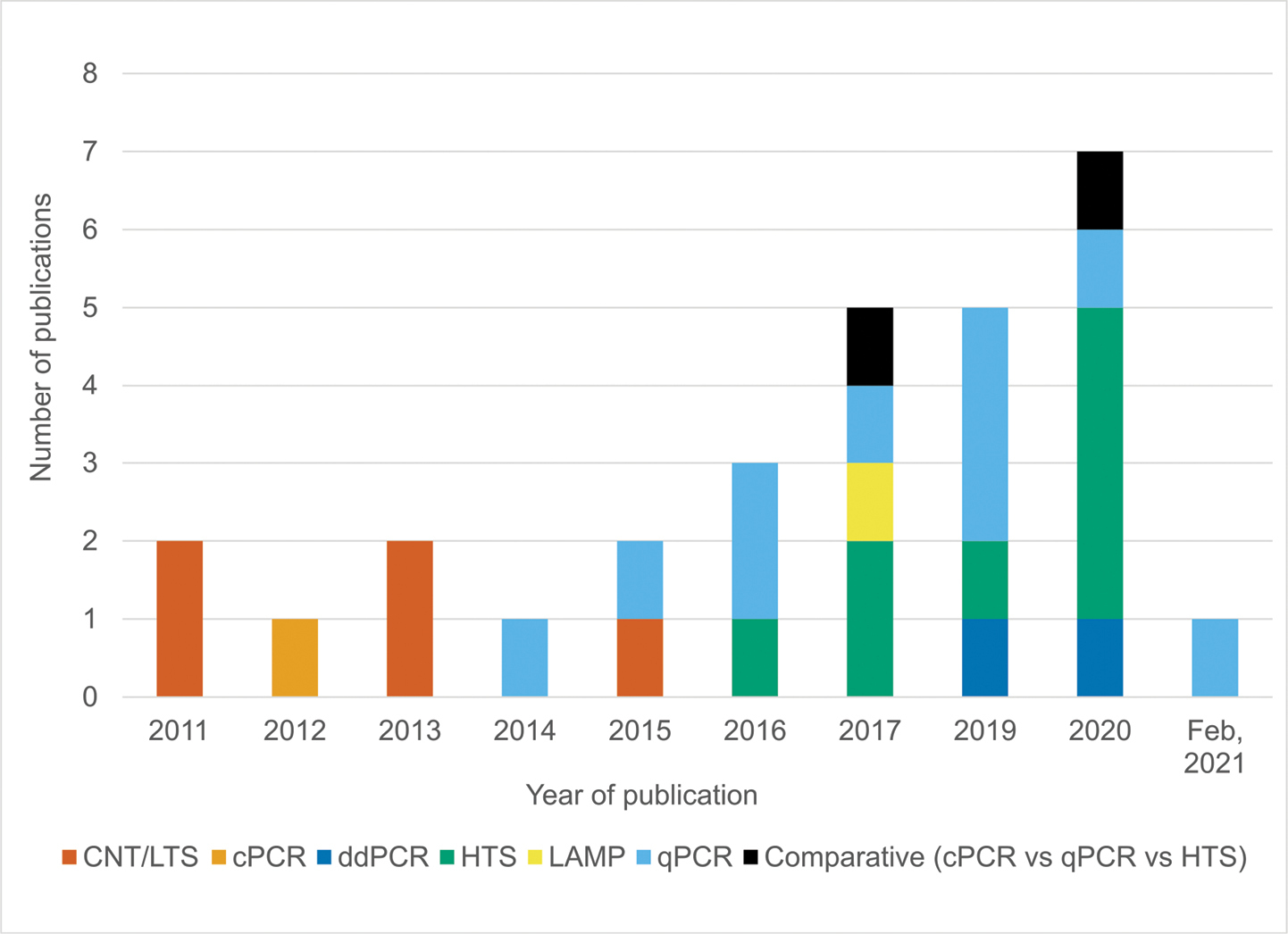
Evolution of quagga mussel and zebra mussel (QM-ZM) environmental DNA methods through time. Numerous technologies have been used to amplify and detect the DNA of QM-ZM contained within environmental samples. Technology types include nanoparticle-based methods (i.e. carbon nanotube or light transmission spectroscopy, CNT/LTS), conventional PCR (cPCR), droplet digital PCR (ddPCR), high-throughput sequencing (HTS), loop-mediated amplification (loop-mediated amplification), quantitative PCR (qPCR) and comparative methods. Here, we can see that methods have evolved over time, with qPCR and HTS currently dominating the field and with ddPCR emerging. |