|
||
|
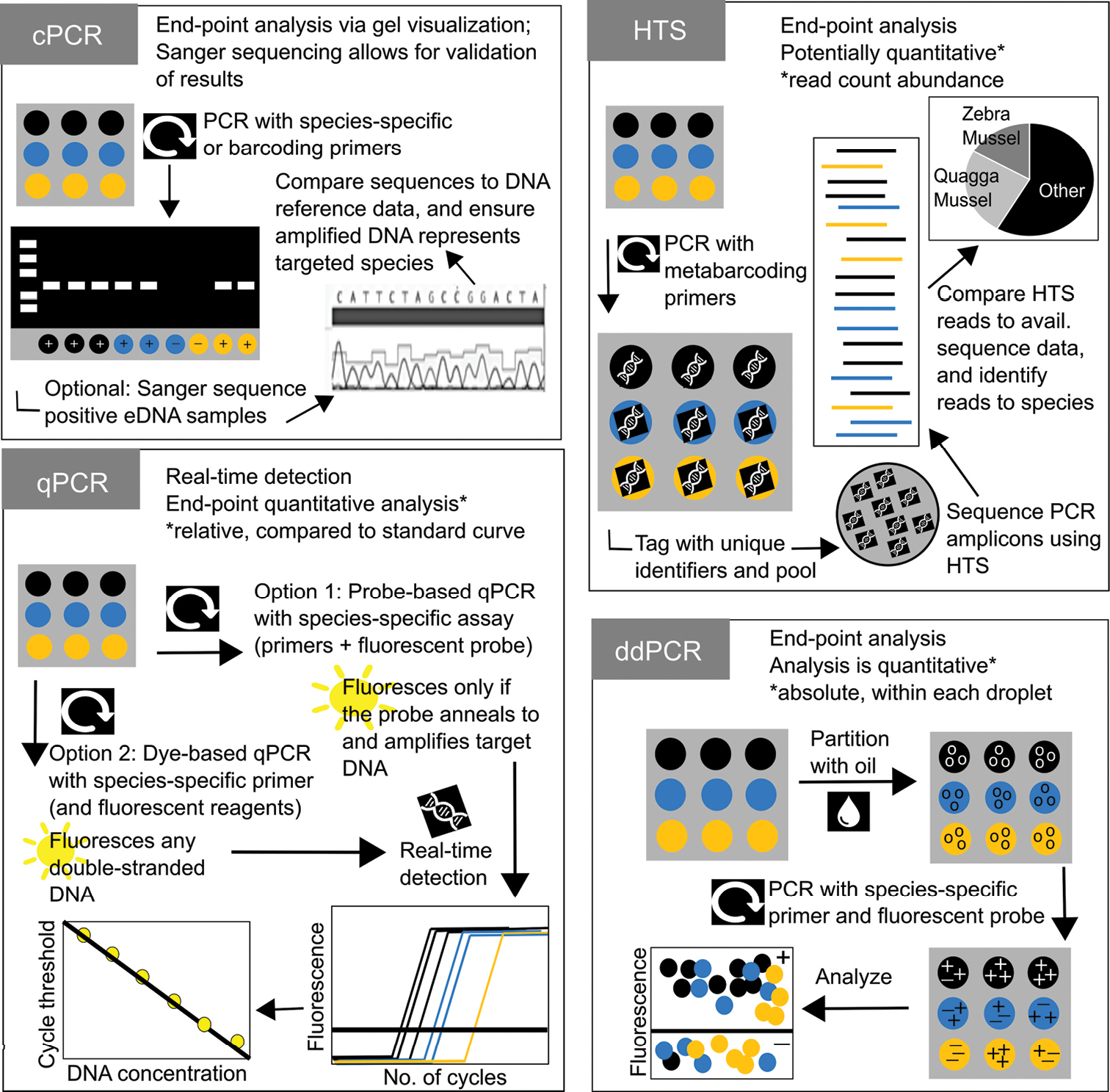
Detailed descriptions of common and emerging quagga mussel and zebra mussel (QM-ZM) environmental DNA (eDNA) amplification strategies. Quantitative PCR (qPCR) and high-throughput sequencing (HTS) represent the most commonly used technologies in quagga and zebra mussel eDNA studies. Droplet digital PCR (ddPCR), an advanced form of qPCR, is an emerging technique with popularity likely to increase due to its high tolerance of PCR inhibitors, improved quantification and observed sensitivity. Here, we detail the specifics of each technique, highlighting how detection and quantification occurs with each. Colours represent three hypothetical environmental DNA (eDNA) samples, at three technical (i.e. lab, amplification) replicates. Positive symbols represent eDNA detection. Negative symbols represent no eDNA detection. Conventional PCR (cPCR) is a foundational technology which gave rise to the other amplification strategies. It is no longer a common eDNA approach (due to low sensitivity), but we include it here for comparative purposes. |