|
||
|
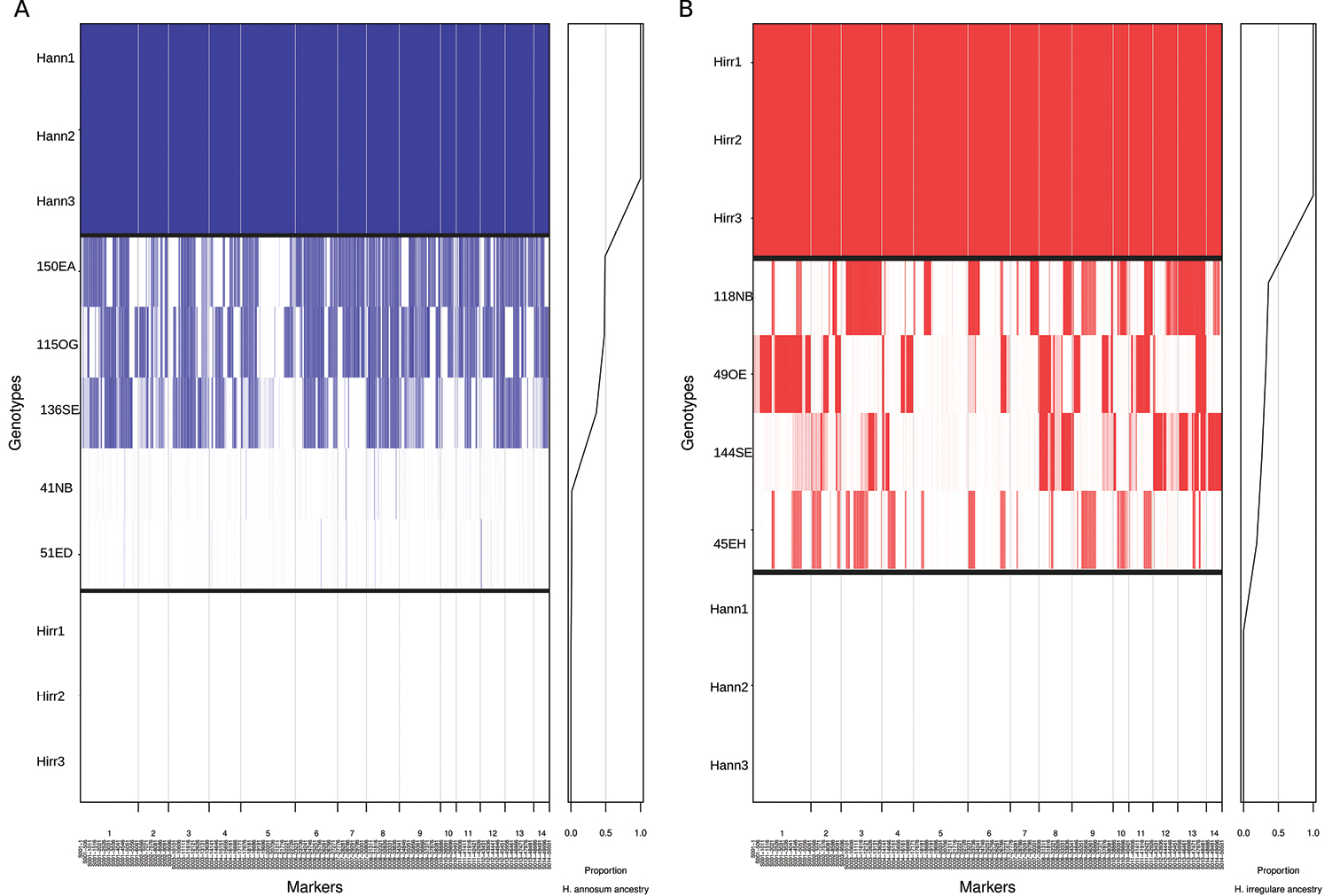
Analysis of introgressed blocks from native to invasive species (A) and from invasive to native species (B) by INTROGRESS. The dataset of used SNPs (markers) are ordered in x-axis based on genomic locations. In (A), Blue rectangles represent marker assigned to H. annosum. Plot on the right represents the fraction of the hybrid genome inherited from H. irregulare for each genotype defined as hybrid index. Three pure genotypes of H. annosum (on the top) are also included in the analysis. In (B), red rectangles represent marker assigned to H. irregulare. Plot on the right represents the fraction of the hybrid genome inherited from H. irregulare for each genotype defined as hybrid index. Three pure genotypes of H. irregulare (on the top) are also included. |