Citation: Moralez-Silva E, Del Lama SN (2014) Colonization of Brazil by the cattle egret (Bubulcus ibis) revealed by mitochondrial DNA. In: Capdevila-Argüelles L, Zilletti B (Eds) Proceedings of 7th NEOBIOTA conference, Pontevedra, Spain. NeoBiota 21: 49–63. doi: 10.3897/neobiota.21.4966
The cattle egret (Bubulcus ibis) has recently colonized Brazil. This process offers an excellent opportunity for the study of colonization and dispersal patterns across extensive areas by non-native birds. The aims of the present investigation were a) to determine the genetic diversity of the cattle egret in Brazil and Africa, b) evaluate genetic differentiation between populations in different regions of Brazil and Africa, and c) detect genetic signs of demographic expansion in these two areas. Mitochondrial DNA (mtDNA) Control Region (CR) sequences were obtained from 112 cattle egrets in four Brazilian and four African (Kenya, Ghana and Nigeria) populations. Genetic diversity (H, h, θs) and population structure (AMOVA, Fst) were assessed and the populations were tested for signs of recent demographic expansion. A total of 35 haplotypes were found: 22 exclusive to Africa, 10 exclusive to Brazil and three shared by both samples. The degree of genetic diversity, determined by mtDNA analysis, was similar between Brazil and Africa, demonstrating that the successful colonization of the non-native area occurred with no significant loss of diversity. The pairwise Fst values among the Brazilian and African populations were all significantly different. The population in southern Brazilian exhibited the lowest degree of differentiation with respect to the African population, followed by the southeastern and northeastern populations of the country. The genetic differentiation data suggest that the colonization of Brazil by the cattle egret began in the southern region and expanded to the southeastern and northeastern regions of the country. This genetic differentiation pattern is in accordance with the higher number of cattle per grazing area in southern Brazil, which may have favored the onset of the successful establishment of the species. The findings indicate that mtDNA genetic diversity was retained during the colonization process and colonization began in the southern region of the country. Moreover, signs of demographic expansion were detected in the African sample.
Cattle egret, colonization, control region, dispersal, diversity
The cattle egret (Bubulcus ibis Linnaeus, 1758) primarily inhabits grassland habitats and forages in close association with grazing animals, such as cattle and other livestock, and it is classified in three subspecies. The subspecies Bubulcus ibis ssp. is native to tropical and subtropical Africa, southern Europe and western Asia (
By approximately 1900, the cattle egret had also expanded its range in Africa (
Novel colonizers can cause problems outside of their native range. While the cattle egret is not currently a threat to native fauna in Brazil throughout most of its geographic distribution, it has the potential to produce adverse effects, as evidenced by its occupation of island environments. For example, in the Fernando de Noronha archipelago, the cattle egret drives adult native seabirds from their nests in breeding colonies (
Successful establishment in a new area by a novel colonizing species is determined by several factors, such as reproductive capacity, growth rate and suitable environmental features (
Genetic tools can provide data that assist in clarifying the process of colonization of non-native areas by alien species. Mitochondrial DNA (mtDNA) has been used in many studies on invasive species to identify historical introduction pathways or determine likely sources of introduced populations (e.g.,
The aim of the present study was to investigate the colonization of Brazil by the cattle egret through the use of mtDNA Control Region (CR) sequences. Inferences are based on four Brazilian samples from different latitudes and four African samples in a more limited geographic area. The cattle egret has considerable dispersal potential and low fidelity to breeding sites (
Two types of biological samples were obtained for the study: blood samples were collected from nestlings in breeding colonies and molted feathers were collected beneath nests in breeding colonies. All sampling sites were reproductive colonies, except the Kankun National Park in Ghana, which is a roosting site. Blood samples (one per nest) were collected from each colony, pooled and each group was considered a single population, i.e., a group of individuals resulting from random interbreeding. The Brazilian samples (N = 51) consisted only of blood samples collected from breeding colonies and all African samples (N = 61) consisted of molted feathers (see Table S1 in Appendix for the complete list of locations sampled). Only feathers in good condition and with no signs of physical degradation were analyzed (
A fragment of 430 bp of the domain I of the CR mtDNA was amplified for 112 cattle egret samples using primers developed in the present study: Ardea L3 (5’-CAC CTA ACA CAA AAC ACA AAC-3’) and BiDIH4 (5’-CTT CAG ATA CCG GTA CTT C-3’). Polymerase chain reactions were conducted in a total volume of 12.5 µL containing 10 ng of template DNA, PCR buffer containing 10 mM Tris-HCl, 50 mM KCl, 20 mM ((NH4)2SO4 and 2 mM MgCl2 (pH 9.0), 0.25 µM of each dNTP, 0.10 µM of each primer and 1 U of Taq DNA Polymerase (Biotools B&M Labs, S.A.). The cycling conditions were follows: denaturation for 5 min at 94 °C; 5 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min; 20 cycles of 94 °C for 30 s, 58 °C for 30 s, decreasing 0.1 °C per cycle; 10 cycles of 94 °C for 30 s, 45 °C for 30 s and 72 °C for 1 min; and a final extension at 72 °C for 10 min. The amplified fragments were sequenced in an ABI Prism 3730 sequencer (Applied Biosystems Inc., California, USA). Sequences were initially aligned with CLUSTALW (
Genetic diversity was examined by calculating the number of haplotypes (H), haplotype diversity (h) and molecular diversity (θ). Genetic diversity estimates were calculated using the ARLEQUIN program v3.5 (
Relationships among haplotypes were inferred through the construction of a statistical parsimony network by the TCS v1.21 program (
Deviation from selective neutrality was tested using Fu’s Fs (
The number of haplotypes was higher in Africa (H = 25) than Brazil (H = 13). Table 1 displays the complete list of haplotypes and frequencies in the populations. Haplotype diversity (h) (mean ± SD) was 0.85 ± 0.04 in Africa and 0.90 ± 0.02 in Brazil. Molecular diversity based on the number of polymorphic sites (θs) was 3.33 ± 1.17 in Africa and 3.11 ± 1.17 in Brazil. The African and Brazilian populations exhibited 16 and 14 polymorphic sites, respectively.
Number of domain I Control Region mtDNA haplotypes evidenced by distinct sequences in cattle egret (Bubulcus ibis) in two areas: A = Africa; B = Brazil.
| S / H | 47 | 69 | 74 | 114 | 117 | 148 | 215 | 242 | 278 | 288 | 290 | 294 | 296 | 316 | 325 | 373 | 375 | 378 | 403 | 413 | B | A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap1 | T | G | G | C | G | A | T | C | T | C | C | A | C | G | C | G | C | C | A | T | 8 | 4 |
| Hap2 | . | . | . | . | . | . | C | T | C | . | . | . | T | . | . | . | T | T | . | . | 11 | - |
| Hap3 | . | . | . | . | . | . | . | T | C | . | . | . | T | . | . | . | T | . | . | . | 4 | 7 |
| Hap4 | . | . | . | . | . | . | . | T | . | . | T | . | T | . | . | . | T | . | . | . | 4 | - |
| Hap5 | . | . | . | . | . | . | . | T | C | . | . | . | T | A | . | . | T | . | . | . | 6 | - |
| Hap6 | . | A | . | . | . | G | . | . | C | . | . | . | T | . | . | . | T | . | . | . | 3 | - |
| Hap7 | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | T | . | . | . | 4 | 2 |
| Hap8 | . | . | . | . | . | . | . | . | . | T | T | . | . | . | . | . | T | . | . | . | 2 | - |
| Hap9 | . | . | . | . | . | . | . | T | C | . | . | G | T | . | . | . | T | . | . | . | 3 | - |
| Hap10 | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | T | . | C | . | 1 | - |
| Hap11 | . | A | . | . | . | . | . | . | . | . | . | . | T | . | . | . | T | . | . | . | 3 | - |
| Hap12 | . | . | . | . | . | . | . | . | C | . | . | G | T | . | . | . | T | . | . | . | 1 | - |
| Hap13 | A | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | 1 | - | |
| Hap14 | . | . | . | . | . | . | . | . | C | . | . | . | T | . | . | . | T | . | . | . | - | 26 |
| Hap15 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | - | 2 |
| Hap16 | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | . | . | . | . | - | 2 | |
| Hap17 | . | . | . | . | . | . | . | T | C | T | . | . | T | . | . | . | T | . | . | . | - | 1 |
| Hap18 | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | T | . | . | . | - | 2 |
| Hap19 | . | . | . | . | . | . | . | T | . | . | . | . | T | . | . | A | T | . | . | . | - | 1 |
| Hap20 | . | . | . | . | . | . | . | . | . | . | . | . | T | . | T | . | T | . | . | . | - | 2 |
| Hap21 | . | . | . | . | A | . | . | . | C | . | . | . | T | . | . | . | T | . | . | . | - | 2 |
| Hap22 | . | A | . | . | . | . | . | . | C | . | . | . | T | . | . | . | T | . | . | . | - | 1 |
| Hap23 | . | . | . | . | . | . | . | . | . | T | . | . | T | . | . | . | T | . | . | . | - | 1 |
| Hap24 | . | . | A | . | . | . | . | T | C | . | . | . | T | . | . | . | T | T | . | . | - | 1 |
| Hap25 | . | . | . | . | . | . | . | . | C | . | T | . | T | . | . | . | T | . | . | . | - | 2 |
| Hap26 | . | . | . | T | . | . | . | T | C | T | . | . | T | . | . | . | T | . | . | . | - | 1 |
| Hap27 | . | . | . | . | . | . | . | . | C | . | . | . | T | . | T | . | T | T | . | . | - | 1 |
| Hap28 | . | . | . | . | . | . | . | . | C | T | . | . | T | . | . | . | T | . | . | . | - | 2 |
| Hap29 | . | . | . | . | . | . | . | . | C | . | . | . | T | . | . | . | T | T | . | . | - | 2 |
| Hap30 | . | . | . | . | . | . | . | . | C | . | . | . | T | . | . | . | . | . | . | . | - | 2 |
| Hap31 | . | . | A | . | . | . | . | . | C | . | . | . | T | A | . | . | T | T | . | . | - | 1 |
| Hap32 | . | . | . | . | . | . | . | T | C | . | . | . | T | . | . | . | . | . | . | . | - | 1 |
| Hap33 | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | T | T | . | . | - | 1 |
| Hap34 | . | . | . | . | . | . | . | T | C | . | . | . | T | . | . | . | T | . | . | C | - | 1 |
| Hap35 | . | . | . | . | . | . | . | T | C | . | . | G | T | . | . | . | T | . | . | . | - | 1 |
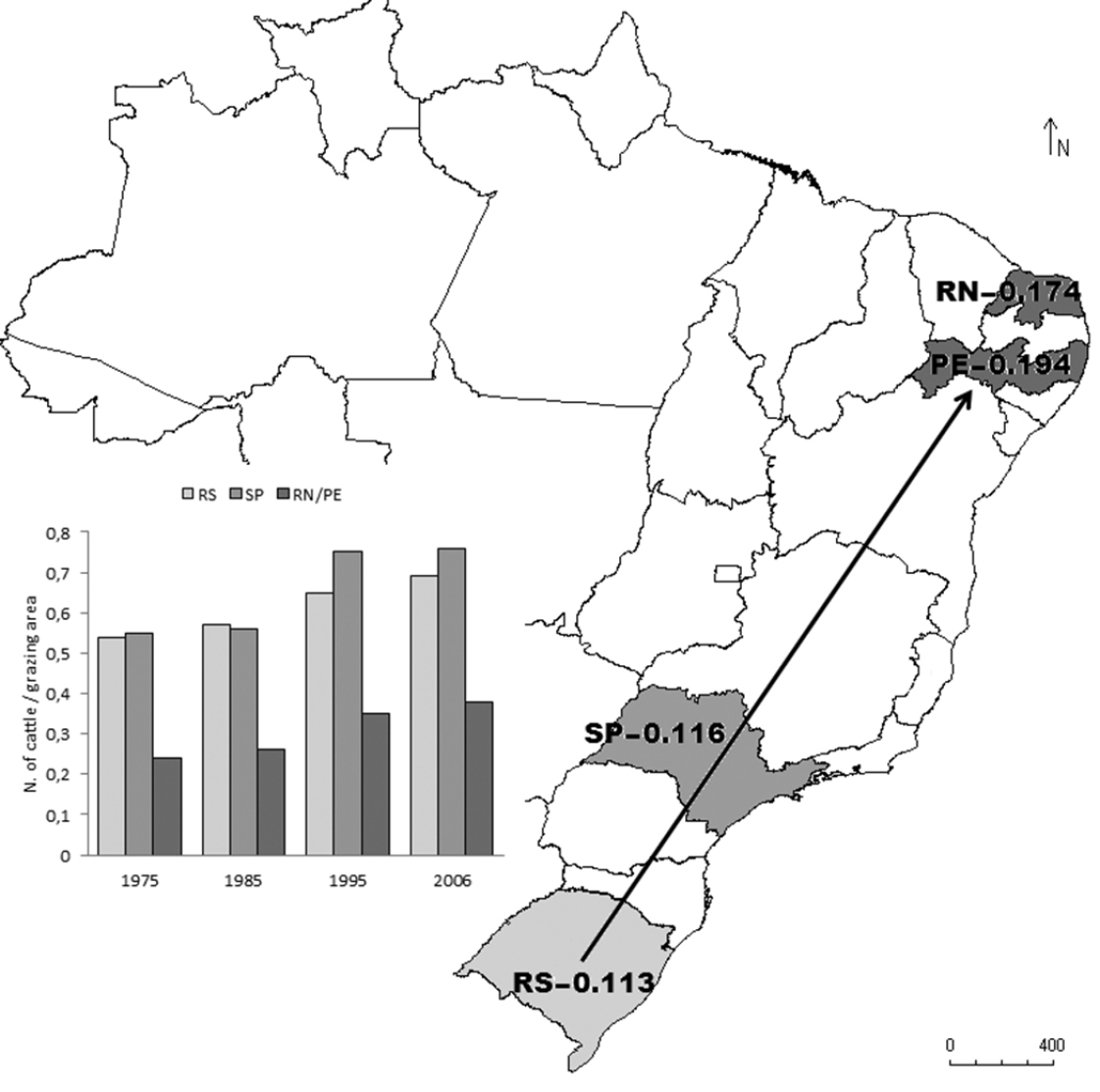
AMOVA revealed that 8.73% of the detected variation was explained by differences between the two populations (African and Brazilian) and the remaining 91.27% was explained by individual differences within populations. The number of haplotypes found in the four Brazilian regions ranged from eight to nine, with no descending cline from the location of the first record of the cattle egret (northern Brazil) to the most distant population sampled (southern Brazil). The pairwise Fst value between the Brazilian and African populations was 0.10. As this value was statistically significant (P< 0.05), pairwise Fst values were calculated among all eight sampling sites (data not shown). Pairwise Fst values involving only Brazilian pairs or African pairs of populations were non-significant, demonstrating no sign of population structuring within each region. The four Brazilian populations were also compared against the entire African population, considering Africa to be one of the probable sources of the Brazilian population, and possible differences in the degree of differentiation between the different Brazilian populations and the entire African population were tested. As expected, significant Fst values were found for all paired comparisons (P < 0.05). The lowest degree of differentiation in relation to the entire African population was found in the southern region of Brazil (state of Rio Grande do Sul [RS]), followed by the southeastern region (state of São Paulo [SP]), and the most differentiated region was northeastern Brazil (states of Rio Grande do Norte [RN] and Pernambuco [PE]) (Fig. 1).
Map of Brazil showing states were cattle egret (Bubulcus ibis) populations were sampled. Levels of differentiation (Fst values) between each Brazilian population and the total African population are shown. Arrow indicates probable path of colonization. Histogram shows the number of cattle per grazing area in four periods (10 years apart) in the states sampled.
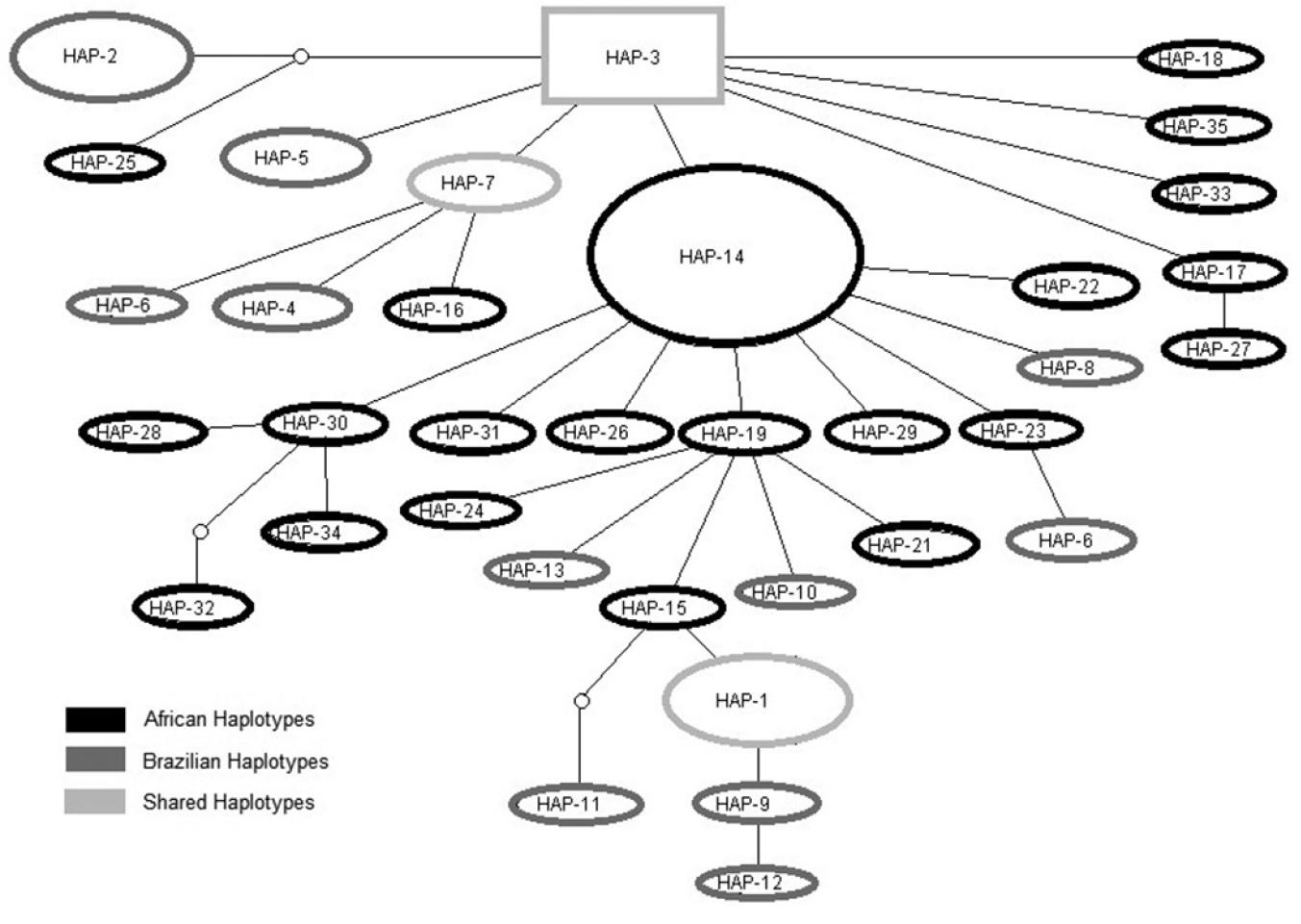
All private Brazilian haplotypes and some of the African haplotypes are located on branch tips in the haplotype network, which includes only three haplotypes shared by both continents (HAP-1, HAP-3 and HAP-7) (Fig. 2). HAP-1 is shared among all Brazilian populations and Ghana. HAP-3 is shared among all African populations and three Brazilian populations (RN, SP and RS). HAP-7 is shared among three Brazilian populations (RN, PE and RS) and Ghana.
Haplotype network and relationships among 35 haplotypes of the domain I CR mtDNA region found in cattle egret (Bubulcus ibis) samples from Brazil and Africa. Areas of shapes are proportional to number of individuals sharing each haplotype.
Neutrality tests were performed assuming that the domain I sequence of the mtDNA is not under selection. Deviations from neutrality in these tests were considered to be indicative of demographic expansion (Table 2). Fu’s Fs and R2 tests revealed significant genetic signs of demographic expansion for the African population. The observed mismatch distribution curves displayed a unimodal shape for African and Brazilian populations and the SSD values were non-significant, indicating that the observed curves did not differ significantly from the population expansion model.
Results of neutrality tests performed for Brazilian and African populations of cattle egret (Bubulcus ibis).
| Population | Tajima’s D | Fu and Li’s D* | Fu and Li’s F* | Fu’s Fs | R2 |
|---|---|---|---|---|---|
| Brazil | 0.45 | 0.57 | 0.63 | -1.50 | 0.10 |
| Africa | -1.28 | -1.25 | -1.50 | -22.25 |
0.10 |
# significant values
Understanding the colonization of the Americas by the cattle egret is a challenging task due to the lack of sufficient information and reports on entrance time, locality, number of events, and propagule pressure. Comparisons between native and non-native populations can provide a ‘natural’ experimental approach to clarify the biological and environmental factors that may contribute to range expansion and/or adaptation to climate change, and to reveal mechanisms by which organisms respond to novel ecological and environmental pressures (
The present study is based solely on results from the analysis of mtDNA, which is maternally inherited, and included a limited sample from the African continent. This analysis revealed no decrease in the level of diversity in the Brazilian samples in relation to the African samples. Indeed, haplotype diversity (h) and molecular diversity (θs) were very similar between the African and Brazilian samples. According to
The majority of mtDNA CR haplotypes were exclusive to either Brazil or Africa, with only three of the total 35 haplotypes shared between both areas. The admixture of Brazilian and African haplotypes could be due to retained ancestral polymorphism in these populations, which is expected for the recent colonization of a new area, such as in the case of the cattle egret in the Americas. The ancestral haplotype (HAP-3) was the fourth most frequent and it was found in both Brazil and Africa, as expected. The most common haplotypes in both the African and Brazilian populations were derived by one or two mutational steps from this ancestral haplotype. It therefore seems that, among the haplotypes sampled, HAP-3 was the first-established lineage, giving rise to other haplotypes, which increased in number with the expansion of the species. All Brazilian haplotypes are on branch tips in the haplotype network. This pattern does not mean that they emerged in the history of Brazilian populations due to the lack of sufficient time for the appearance of new mutations, but suggests that these haplotypes emerged more recently in the evolutive history of these sequences. African haplotypes on branch tips reveal that the African populations sampled are in a geographic range where expansion on the native continent also occurred. The detection of shared haplotypes (among all Brazilian samples and mostly Ghana) and the fact that the ancestral haplotype is present in both regions support a common origin for the African and Brazilian populations sampled.
Genetic differentiation between populations from Brazil and Africa was evidenced by statistically significant pairwise Fst values obtained for all paired comparisons in the two areas. Despite the fact that the African samples were not representative of all regions of the continent, the supposition was that the least differentiated population sampled in the non-native area in relation to the African samples would be more similar to the founder population. The results of the mtDNA analysis of genetic differentiation indicated that Brazil was first colonized in the southern region (RS population) (Figure 1). The first record of a breeding colony of cattle egrets in Brazil was on Marajó Island in the northern region of the country (
All pairwise Fst values between the Brazilian and African populations were statistically significant, as can be seen in Figure 1. This suggests a certain degree of constraint in gene flow. On the other hand, the partial admixture of Brazilian and African haplotypes detected in the haplotype network can be interpreted as a sign of gene flow between these two areas. The possibility of continuing migration to South America from different (non-sampled) regions of Africa cannot be ruled out, since a limited sample from Africa was included in this study.
The present findings revealed no signs of demographic expansion in Brazil, as expected for a recent colonization process. However, Fs and R2 values and the unimodal mismatch curves revealed significant genetic signs of demographic expansion in the African samples. According to
The cattle egret has retained most of the mtDNA genetic diversity during the colonization process in Brazil. Genetic signs of demographic expansion were detected in the African sample. The genetic differentiation analysis shows that this species began colonization in the southern region of the country and expanded in a northeasterly direction to the southeastern and northeastern regions. This dispersal pattern is supported by the environmental characteristics and larger amount of cattle raising and pasture areas in the south region in comparison to the two other regions sampled. Future studies involving a greater number of samples in its native range and the inclusion of an analysis of nuclear genes will provide a more complete scenario of the proposed colonization process of the cattle egret in Brazil.
We acknowledge the comments and suggestions made by the reviewers, which greatly contributed to improving the quality of this manuscript. EMS acknowledges support from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-Brazil, 870195/1997-2) for a Ph.D grant. SNDL received funds from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP-Brazil, 2010/50406-5). The permit for the collection of samples was provided by the Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio- Number 20295-2) and the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais (IBAMA). Permits for the importation of samples were provided by the Instituto Brasileiro do Meio Ambiente (IBAMA– Number 11BR007199/DF, 11BR007010/DF). The authors are grateful to Roberta Rodrigues from CEMAVE, Jacinta Abalaka and Dr. Shiiwa Manu from A. P. Leventis Ornithological Research Institute (APLORI), Daryl Bosu from NGO A Rocha Ghana for collecting samples, Mr. Scherezino Scherer (IBAMA) for helping with the field expedition in RS and Carlos Congrains Castillo for help with data analysis.
Number of samples and locations in Brazil and Africa.
| Location name | Number of samples | Geographic coordinates | |
|---|---|---|---|
| BRAZIL | Pernambuco state - PE | 13 | 8°52'S, 36°28'W |
| Rio Grande do Norte state - RN | 12 | 5°37'S, 36°52'W | |
| São Paulo state - SP | 13 | 22°30'S, 47°35'W | |
| Rio Grande do Sul state – RS | 13 | 30°01'S, 51°31'W | |
| AFRICA | Nigeria | 11 | 9°58'N, 8°53'E |
| Kenya | 20 | 1°19'S, 36°51'E | |
| Kakun National Park - Ghana | 15 | 05°32' N, 01°13W | |
| Korle Lagoon, Accra city - Ghana | 15 | 09°05'N, 01°49W |