Review Article |
|
Corresponding author: Claudia Romeo ( claudia.romeo@sund.ku.dk ) Academic editor: Jonathan M. Jeschke
© 2025 Claudia Romeo, Elsa Brenner, Lucas A. Wauters, Antton Alberdi.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Romeo C, Brenner E, Wauters LA, Alberdi A (2025) The role of microbiomes in animal invasions: a scoping review. NeoBiota 98: 335-360. https://doi.org/10.3897/neobiota.98.145939
|
Abstract
Despite increasing evidence for the contribution of microbiomes to host fitness and evolution, their role in the adaptation and successful establishment of invasive animal species remains underexplored. Animal microbiomes can mediate key host phenotypic traits such as energy metabolism, detoxification and disease resistance. Therefore, harbouring a highly functional microbiome may be advantageous in the context of invasion, where small host populations must rapidly adapt to new environmental conditions. We conducted a scoping review of studies focusing on microbiomes and animal invasions to explore the extent and nature of research efforts on this topic and to identify general patterns that may help elucidate the relationship between host microbial communities and invasiveness. The analysis of 147 articles published between 2006 and 2024 showed a steady increase in the research output on the topic, in parallel with growing interest in biological invasions and technical and theoretical advances in microbiome research. However, the application of new analytical approaches that go beyond taxonomic characterisation remains limited and the research output is still heavily biased towards invasive invertebrates. Although most of the reviewed research was descriptive, a more detailed assessment of a subset of 43 studies using a comparative design revealed some recurring patterns. Host microbiomes in the introduction range tend to diverge from those in the native range, but invasive populations generally retain a core of microorganisms involved in key phenotypic traits such as disease resistance. Studies that have examined the microbiomes of invasive species along their invasion pathway highlight how stochastic events, propagule pressure and population mixing are relevant drivers of microbial community assembly during introductions. Comparisons of the microbiomes of invasive species and co-occurring, outcompeted native species often suggest that some of the observed phenotypic differences driving their interactions are microbiome-mediated. However, to date, only a handful of studies have been able to establish the mechanistic link between microbiomes and host invasiveness using an experimental design. While observational studies remain valuable at this early stage, we advocate for a wider use of novel technologies and experimental approaches to generate robust functional and mechanistic information that will strengthen their inferential value. As more system-specific studies become available, meta-analytical approaches may allow us to uncover broader eco-evolutionary patterns and ultimately elucidate the role of microbiomes in animal invasions.
Key words:
Adaptation, alien species, biological invasions, invasiveness, invasive species, metagenomics, microbial community, microbiota
Introduction
Most animals harbour complex communities of microorganisms, the animal-associated microbiota (
Over fifteen years ago,
In the last decades, evidence for a role of the microbiome in animal fitness, adaptation and evolution has been increasing (
Biological invasions occur when a — usually small — number of individuals are translocated by humans to a new area outside their natural range, where they establish a viable population and spread away from the point of introduction (
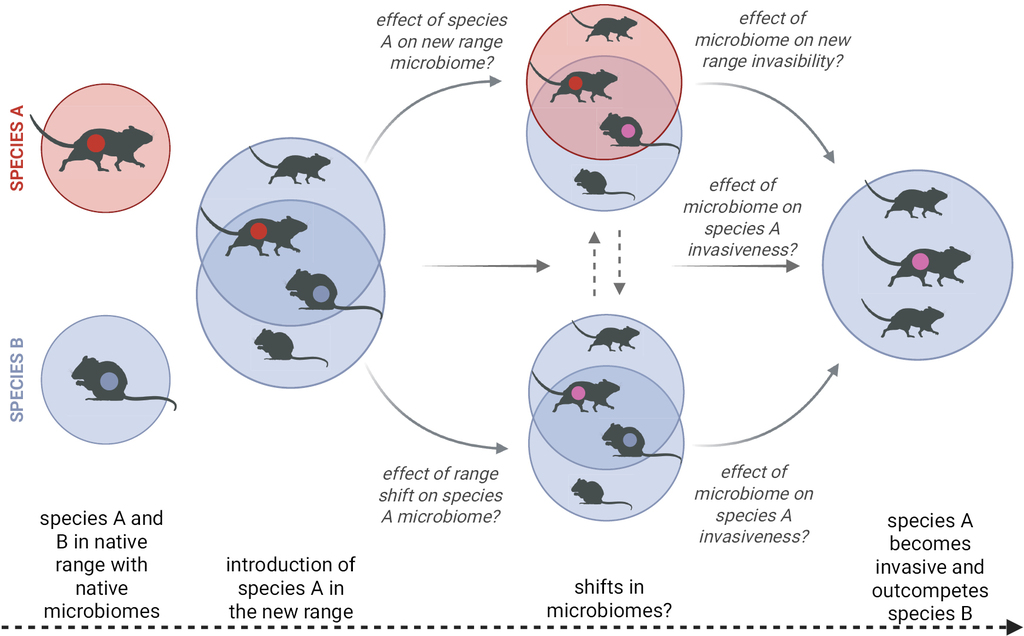
However, as yet, the link between microbiomes and animal invasions remains somewhat underexplored. It is important to note that the interaction between microbiomes and invasions is likely to go both ways and disentangling causality can be challenging. As illustrated in Fig.
Microbiome dynamics during invasions. Illustration of the potential two-way interactions between animal microbiomes and the invasion process. Created in BioRender (https://BioRender.com/d20m325).
For instance, some animal species — especially aquatic or soil invertebrates — have the capability to alter the environmental microbiota surrounding them via microbiome excretion and/or their mechanical or chemical actions. This can increase habitat invasibility and greatly affect local communities, facilitating the establishment and spread of invaders (
To collate up-to-date evidence on this topic, extend the analysis to vertebrates and highlight current knowledge gaps and research perspectives, we conducted a scoping review (
Literature search and analysis
The literature search was conducted in May 2024 using both Web of Science and Scopus platforms and following the PRISMA protocol (
- the study included at least one invasive animal as a focal species (hence, studies not pertaining to invasion biology or focussing on plant or bacterial invasions were excluded); and
- the study investigated the focal species’ microbiota and/or the impact of the focal species on the microbiota of either a native counterpart or the local environment; and
- the study examined the microbiota from a community ecology perspective. Hence, studies that only focussed on the pathobiome or on specific bacteria as a means for biocontrol were excluded.
Concerning the first criterion, although the term “invasive” was sometimes used more loosely in the retrieved studies, we included only studies where the translocation of the focal species to a new geographic range was human-mediated, either intentionally or unintentionally.
Deduplication and title and abstract screening were conducted using Rayyan (
The analysis of included literature was conducted in two stages. First, each of the 147 articles was classified using the descriptors listed in Table
List of descriptors used to classify the 147 articles included in the present review. A detailed explanation of the study design descriptors can be found in Suppl. material
| Article type | Animal group | Animal taxon | Target species | Geographic design | Study design | Microbiota location | Method |
|---|---|---|---|---|---|---|---|
| Research | Invertebrate | Arthropod | Only invasive sp. | Only native range | Comparative: range and host | Only environment | Amplicon-based sequencing |
| Review | Vertebrate | Invertebrate (Other) | Only native sp. | Only introduction range | Comparative: range | Gut | Shotgun metagenomics |
| Amphibian | Comparative | Only invasion wave | Comparative: host species | Multiple organs/tissues | Other | ||
| Bird | Comparative | Comparative: IASa traits | Oral | ||||
| Fish | Descriptive: IASa | Skin/Exoskeleton | |||||
| Mammal | Descriptive: impact | Whole specimen | |||||
| Reptile |
Second, we selected the subset of studies that focussed specifically on host-associated microbiomes and used a comparative design to address the hypothesis of a microbial contribution to invasiveness. This led to the identification of 43 articles either comparing: i) the microbiota of an invasive versus a native species, ii) the microbiota of an invasive species in the introduction versus the native range or iii) the microbiota of an invasive species along the invasion wave. From this subset of selected papers, we identified the main recurring patterns and will report a qualitative synthesis of their key findings.
Trends in animal invasions and microbiome research
The 147 papers which met all the inclusion criteria spanned 2006-May 2024, with a median publication year of 2020, demonstrating a steady increase in output over time (Fig.
Trends in microbiomes-animal invasions research. Trends in the research output on microbiomes and animal invasions a number of articles published by year, methods used to characterise the microbial community and study design (the black line indicates the number of comparative studies); and b number of articles published by invasive host taxon and location of the target microbial community.
The classification of the 140 research articles by the methodology used to characterise the microbiota yielded that the vast majority (87.9%, n = 123) of papers applied a targeted amplicon sequencing-based approach (16S rRNA for prokaryotes, 18S rRNA for eukaryotes, ITS for fungi or a combination of these). The remaining 12.1% used shotgun metagenomics (5.7%, n = 8), while the rest relied on other approaches (6.4%, n = 9), such as Restriction Fragment Length Polymorphisms (RFLP), microscopy or staining (Fig.
Most research articles focused on invasive arthropods (44.3%, n = 62) or other invertebrates (32.1%, n = 44), whereas vertebrates were the focus of 23.6% (n = 33) of the articles (Fig.
Research also varied in terms of which of the host’s microbial communities was studied (Fig.
In terms of design, 39.9% (n = 56) of the research articles were descriptive, with a primary focus on characterising the composition of an invasive species’ microbiome or its impact on the surrounding environment, while the remaining 60.1% (n = 84) of research papers included a comparative aspect. Earlier studies were more frequently descriptive in nature, while, from 2018 onwards, there is an increase in the number of more complex, comparative study designs that try to infer the role of the microbiome in the invasive species’ adaptation (Fig.
Patterns in animal invasions and the microbiome
We used the subset of the 43 comparative research papers for a more in-depth analysis to gain further insight into whether the microbiome is a driver or facilitator of invasiveness. The articles included in the subset of comparative studies either compared the microbiome of the invasive species to a native counterpart (17 articles) or the microbiome of the invasive species across its native and introduction range (17 articles) or along the invasion wave (6 articles). Three articles compared the microbiome of the invasive species both against the competing native species and across ranges. Most of these studies still targeted invertebrate hosts (26 articles), but vertebrates were relatively well represented (14 articles). We observed that a few invasive species-microbiome systems have been explored more in depth through multiple comparative studies (listed in Table
List of the invasive animal-microbiome systems assessed by multiple comparative studies with corresponding references.
| Invasive host species | References |
|---|---|
| Asian tiger mosquito (Aedes albopictus) | ( |
| Common wasp (Vespula vulgaris) | ( |
| Oriental fruit fly (Bactrocera dorsalis) | ( |
| Warty comb jelly (Mnemiopsis leidyi) | ( |
| Signal crayfish (Pacifastacus leniusculus) | ( |
| Lionfish species complex (Pterois volitans/P. miles) | ( |
| Red-eared slider (Trachemys scripta elegans) | ( |
| Cane toad (Rhinella marina) | ( |
The microbiome across geographic ranges
A prominent question when addressing biological invasions and microbiomes is certainly the fate of an invasive host’s microbial community after its establishment in a new range. In most cases and across a range of diverse host taxa, microbiomes in the invaded range were found to be significantly distinct from those in the native range (Diptera:
Microbiomes across ranges. The microbiome of invasive populations is often distinct from that of populations in the native range, but they typically retain a core of microbial species involved in modulating key phenotypic traits. Created in BioRender (https://BioRender.com/a68e550).
There are, however, a few exceptions to this pattern: treehoppers Stictocephala bisonia (
Regarding the diversity of microbiomes across ranges, several of the comparative studies found that individuals from invasive populations had, on average, higher microbial richness (
The microbiome across competing species
Some further insight into diversity patterns and invasions comes from those studies that compared the microbiota of an invasive species to that of some ecologically similar, co-occurring native species. In many cases, such studies found that the invader harboured a microbial community that was taxonomically and functionally more diverse compared to the native species (
Microbiomes across species. The microbiome of invasive species often has a higher functional potential than that of co-occurring, outcompeted native species. Created in BioRender (https://BioRender.com/v46k439).
Regardless of diversity patterns and similar to what emerged from across-range comparisons, in most cases, the microbiome of the invasive species was clearly distinct from that of co-occurring native species, even when they were phylogenetically very close and/or ecologically very similar (
Drivers of microbiome assembly during invasions
The microbiome is an assemblage of species acquired by the host through a combination of vertical transfer from its parents and horizontal transfer from other co-occurring organisms and the environment (
Studies that analysed variation in the structure and composition of microbiomes along the invasion path of a species can offer further insight into microbiome dynamics during invasions. For instance, Argentine ants (Linepithema humile) were introduced through serial jumps from Argentina to the USA, to Australia and finally to New Zealand and their microbial communities show a progressive decrease in richness and in the relative abundance of core taxa along this path, mirroring a series of bottlenecks in the host population (
Decoupling stochastic processes from adaptive shifts in the microbiota can be challenging.
As seen with other, non-invasive species (
Finally, it must be considered that several vertebrate invasions result from accidental or intentional release from captivity and captivity is another factor that can strongly alter microbial assemblages (
The microbiome as a driver of invasiveness
Although most of the research included in the present review is observational, some recent studies explored the mechanistic link between microbiome composition and invasiveness traits through experimental set-ups.
In a similar experiment,
Another experiment addressing the role of microbiomes in invasions concerns the fall webworm (Hyphantria cunea), a globally invasive North American moth. During the last decades, the species has spread throughout China’s temperate regions and it is now expanding further south, into subtropical areas.
As mentioned before, repeated introductions followed by population mixing could promote increased diversity of microbial communities and prove beneficial for host fitness. Populations of the invasive oriental fruit fly (B. dorsalis) in northern China appear to be hybrids of lineages originating from different biogeographical regions following multiple introductions (
Knowledge gaps, best practices and research priorities
Our review of the existing literature shows that most of the published studies are still descriptive, that technical and analytical methods are far from standardised and that some host taxa are still poorly represented, hindering the possibility of conducting robust quantitative meta-analyses. However, publication trends suggest that interest in the topic is steadily growing and we are confident that more data will soon become available, allowing for meta-analytical approaches that will enable researchers to address broader eco-evolutionary questions regarding the role of animal-associated microbiomes in invasions.
For instance, contrasting diversity patterns emerged from both across ranges and across species comparisons, but the number of studies is still too limited to identify any consistent associations with, for example, host phylogeny. Another interesting question to address would be whether it is more advantageous for an invading species to have a more plastic or conversely a more resistant microbial community. From a slightly different perspective, one could also ask whether some bacterial taxa — or functions — might be more beneficial to conserve — or acquire — than others. It is likely that the answers to such questions would be highly dependent on the host species, the invasion context and the specific functional role of the different microbial taxa, but as more data become available, meta-analyses could potentially reveal broader underlying patterns related to the characteristics of the invaded habitats or the phylogeny and/or niche specialisation of the host or microbial taxa.
Further system-specific research is, therefore, needed to enable researchers to address these broader questions, but it is important that future studies adhere to some common standards in order to be comparable and have inferential value. For instance, our review highlights that the vast majority of comparative studies are still observational in nature, echoing the findings of a recent systematic review on microbe-driven adaptation in wild vertebrates (
Nevertheless, we argue that, at this early stage, comparative, observational studies are still valuable to shed light on whether animal microbiomes may be relevant drivers of invasiveness in any way. In such a high-dimensional and complex system as the host and its microbiome, correlative studies can help to sort out potentially influential patterns that can later be addressed by an experimental approach to prove causality and determine its direction (
However, two requirements are essential for such correlative studies to be meaningful and have some inferential value: first, the use of an appropriate sampling design and second, the generation of robust functional information alongside taxonomic data. Wild animals’ microbiomes usually show high intra- and inter-individual variation; hence, working at the appropriate spatial and temporal scales, sampling multiple populations, as well as choosing the right microbial taxonomic resolution are fundamental to avoid sampling artefacts (
In terms of functional inference, we advocate for a more widespread use of shotgun metagenomics, since reliable and complete functional information is critical for drawing conclusions about observed microbiome shifts or differentially abundant microbial taxa (
Conclusions
Our review of the existing literature shows that the attention of the scientific community to the role of the microbiome as a potential driver of animal invasions has steadily increased over time, but research is still taxonomically biased and mostly observational in nature. The analysis of the subset of comparative studies shows that, in most systems, the host microbiome undergoes relevant changes during the introduction process and many of these shifts appear to have some adaptive value. Several studies also highlight the importance of stochastic processes in determining the post-invasion microbial community. However, to date, only a handful of experimental studies have demonstrated the mechanistic link between the microbiota and invasiveness in an animal species. More such studies are needed to elucidate whether adaptive shifts in microbial communities following invasion are a common occurrence. We believe that observational studies remain valuable, but only when combined with a robust sampling design and strengthened by measures of host fitness and the adoption of new analytical approaches that allow for more robust functional inference. As more complete, system-specific studies become available, meta-analytic approaches will allow researchers to compare the dynamics of microbial communities across multiple invasive species and ecosystems and, potentially, uncover broader eco-evolutionary patterns related to the role of microbiomes in animal invasions.
Acknowledgements
We thank Amalia Bogri for producing some of the illustrations included in Figs
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
CR was funded by the European Union through an MSCA Postdoctoral Fellowship (HORIZON-MSCA-2021-PF-01; Grant ID: 101066225). AA acknowledges the Danish National Research Foundation through the grant DNRF143.
Author contributions
Conceptualisation: CR, AA. Data curation: CR. Formal analysis: EB. Investigation: CR, EB. Methodology: CR, EB. Supervision: CR, AA. Validation: LAW, AA. Visualisation: CR, EB. Writing - original draft: CR. Writing - review and editing: CR, EB, LAW, AA.
Author ORCIDs
Claudia Romeo https://orcid.org/0000-0002-5964-2847
Elsa Brenner https://orcid.org/0009-0003-1313-4593
Lucas A. Wauters https://orcid.org/0000-0002-4871-5035
Antton Alberdi https://orcid.org/0000-0002-2875-6446
Data availability
No new data were analysed or generated for this study. The full list of articles included in the review is provided as Suppl. material
References
- Abarca JG, Zuniga I, Ortiz-Morales G, Lugo A, Viquez-Cervilla M, Rodriguez-Hernandez N, Vázquez-Sánchez F, Murillo-Cruz C, Torres-Rivera EA, Pinto-Tomás AA, Godoy-Vitorino F (2018) Characterization of the skin microbiota of the cane toad Rhinella cf. marina in Puerto Rico and Costa Rica. Frontiers in Microbiology 8: 2624. https://doi.org/10.3389/fmicb.2017.02624
- Aizpurua O, Dunn RR, Hansen LH, Gilbert MTP, Alberdi A (2023) Field and laboratory guidelines for reliable bioinformatic and statistical analysis of bacterial shotgun metagenomic data. Critical Reviews in Biotechnology 0: 1–19. https://doi.org/10.1080/07388551.2023.2254933
- Aketarawong N, Bonizzoni M, Thanaphum S, Gomulski LM, Gasperi G, Malacrida AR, Gugliemino CR (2007) Inferences on the population structure and colonization process of the invasive oriental fruit fly, Bactrocera dorsalis (Hendel). Molecular Ecology 16: 3522–3532. https://doi.org/10.1111/j.1365-294X.2007.03409.x
- Alberdi A, Aizpurua O, Bohmann K, Zepeda-Mendoza ML, Gilbert MTP (2016) Do Vertebrate Gut Metagenomes Confer Rapid Ecological Adaptation? Trends in Ecology & Evolution 31: 689–699. https://doi.org/10.1016/j.tree.2016.06.008
- Alberdi A, Martin Bideguren G, Aizpurua O (2021) Diversity and compositional changes in the gut microbiota of wild and captive vertebrates: A meta-analysis. Scientific Reports 11: 22660. https://doi.org/10.1038/s41598-021-02015-6
- Arias M, Hartle-Mougiou K, Taboada S, Vogler A, Riesgo A, Elfekih S (2022) Unveiling biogeographical patterns in the worldwide distributed Ceratitis capitata (medfly) using population genomics and microbiome composition. Molecular Ecology 31: 4866–4883. https://doi.org/10.1111/mec.16616
- Arksey H, O’Malley L (2005) Scoping studies: Towards a methodological framework. International Journal of Social Research Methodology 8: 19–32. https://doi.org/10.1080/1364557032000119616
- Azzurro E, Nourigat M, Cohn F, Ben Souissi J, Bernardi G (2022) Right out of the gate: The genomics of Lessepsian invaders in the vicinity of the Suez Canal. Biological Invasions 24: 1117–1130. https://doi.org/10.1007/s10530-021-02704-3
- Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, Chen X, Cocolin L, Eversole K, Corral GH, Kazou M, Kinkel L, Lange L, Lima N, Loy A, Macklin JA, Maguin E, Mauchline T, McClure R, Mitter B, Ryan M, Sarand I, Smidt H, Schelkle B, Roume H, Kiran GS, Selvin J, de Souza RSC, van Overbeek L, Singh BK, Wagner M, Walsh A, Sessitsch A, Schloter M (2020) Microbiome definition re-visited: Old concepts and new challenges. Microbiome 8: 103. https://doi.org/10.1186/s40168-020-00875-0
- Blackburn TM, Pyšek P, Bacher S, Carlton JT, Duncan RP, Jarošík V, Wilson JRU, Richardson DM (2011) A proposed unified framework for biological invasions. Trends in Ecology & Evolution 26: 333–339. https://doi.org/10.1016/j.tree.2011.03.023
- Brinker P, Fontaine MC, Beukeboom LW, Falcao Salles J (2019) Host, Symbionts, and the Microbiome: The Missing Tripartite Interaction. Trends in Microbiology 27: 480–488. https://doi.org/10.1016/j.tim.2019.02.002
- Caragata EP, Tikhe CV, Dimopoulos G (2019) Curious entanglements: Interactions between mosquitoes, their microbiota, and arboviruses. Current Opinion in Virology 37: 26–36. https://doi.org/10.1016/j.coviro.2019.05.005
- Catford JA, Jansson R, Nilsson C (2009) Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Diversity & Distributions 15: 22–40. https://doi.org/10.1111/j.1472-4642.2008.00521.x
- Chiarello M, Bucholz J, McCauley M, Vaughn S, Hopper G, Gonzalez I, Atkinson C, Lozier J, Jackson C (2022) Environment and co-occurring native mussel species, but not host genetics, impact the microbiome of a freshwater invasive species (Corbicula fluminea). Frontiers in Microbiology 13: 800061. https://doi.org/10.3389/fmicb.2022.800061
- Christian K, Weitzman C, Rose A, Kaestli M, Gibb K (2018) Ecological patterns in the skin microbiota of frogs from tropical Australia. Ecology and Evolution 8: 10510–10519. https://doi.org/10.1002/ece3.4518
- Coats VC, Rumpho ME (2014) The rhizosphere microbiota of plant invaders: An overview of recent advances in the microbiomics of invasive plants. Frontiers in Microbiology 5: 368. https://doi.org/10.3389/fmicb.2014.00368
- Comizzoli P, Power ML, Bornbusch SL, Muletz-Wolz CR (2021) Interactions between reproductive biology and microbiomes in wild animal species. Animal Microbiome 3: 87. https://doi.org/10.1186/s42523-021-00156-7
- Dallas JW, Warne RW (2023) Captivity and animal microbiomes: Potential roles of microbiota for influencing animal conservation. Microbial Ecology 85: 820–838. https://doi.org/10.1007/s00248-022-01991-0
- Daly EZ, Chabrerie O, Massol F, Facon B, Hess MCM, Tasiemski A, Grandjean F, Chauvat M, Viard F, Forey E, Folcher L, Buisson E, Boivin T, Baltora-Rosset S, Ulmer R, Gibert P, Thiébaut G, Pantel JH, Heger T, Richardson DM, Renault D (2023) A synthesis of biological invasion hypotheses associated with the introduction–naturalisation–invasion continuum. Oikos 2023: e09645. https://doi.org/10.1111/oik.09645
- Davidson GL, Raulo A, Knowles SCL (2020) Identifying microbiome-mediated behaviour in wild vertebrates. Trends in Ecology & Evolution 35: 972–980. https://doi.org/10.1016/j.tree.2020.06.014
- Dearing MD, Kohl KD (2017) Beyond fermentation: Other important services provided to endothermic herbivores by their gut microbiota. Integrative and Comparative Biology 57: 723–731. https://doi.org/10.1093/icb/icx020
- Degregori S, Wang X, Kommala A, Schulhof N, Moradi S, MacDonald A, Eblen K, Jukovich S, Smith E, Kelleher E, Suzuki K, Hall Z, Knight R, Amato KR (2024) Comparative gut microbiome research through the lens of ecology: Theoretical considerations and best practices. Biological Reviews of the Cambridge Philosophical Society 100: 748–763. https://doi.org/10.1111/brv.13161
- Do Y, Park W, Park J, Kim C, Choi M (2023) Gut bacterial diversity in Vespa velutina and implications for potential adaptation in South Korea. Pest Management Science 79(2): 5180–5185. https://doi.org/10.1002/ps.7721
- Douglas AE, Werren JH (2016) Holes in the Hologenome: Why host-microbe symbioses are not holobionts. mBio 7(2): 10. https://doi.org/10.1128/mBio.02099-15. https://doi.org/10.1128/mbio.02099-15
- Dragičević P, Bielen A, Petrić I, Vuk M, Žučko J, Hudina S (2021) Microbiome of the successful freshwater invader, the signal crayfish, and its changes along the invasion range. Microbiology Spectrum 9: e00389-21. https://doi.org/10.1128/Spectrum.00389-21
- Dragičević P, Rosado D, Bielen A, Hudina S (2024) Host-related traits influence the microbial diversity of the invasive signal crayfish Pacifastacus leniusculus. Journal of Invertebrate Pathology 202: 108039. https://doi.org/10.1016/j.jip.2023.108039
- Duguma D, Hall M, Smartt C, Neufeld J (2017) Temporal variations of microbiota associated with the immature stages of two florida culex mosquito vectors. Microbial Ecology 74: 979–989. https://doi.org/10.1007/s00248-017-0988-9
- Erfmeier A (2013) Constraints and release at different scales – The role of adaptation in biological invasions. Basic and Applied Ecology 14: 281–288. https://doi.org/10.1016/j.baae.2013.04.004
- Escalas A, Auguet J, Avouac A, Belmaker J, Dailianis T, Kiflawi M, Pickholtz R, Skouradakis G, Villéger S (2022) Shift and homogenization of gut microbiome during invasion in marine fishes. Animal Microbiome 4: 37. https://doi.org/10.1186/s42523-022-00181-0
- Ferlian O, Eisenhauer N, Aguirrebengoa M, Camara M, Ramirez-Rojas I, Santos F, Tanalgo K, Thakur MP (2018) Invasive earthworms erode soil biodiversity: A meta-analysis. Journal of Animal Ecology 87: 162–172. https://doi.org/10.1111/1365-2656.12746
- Fontaine SS, Kohl KD (2020) Gut microbiota of invasive bullfrog tadpoles responds more rapidly to temperature than a noninvasive congener. Molecular Ecology 29: 2449–2462. https://doi.org/10.1111/mec.15487
- Fontaine SS, Kohl KD (2023) The microbiome buffers tadpole hosts from heat stress: A hologenomic approach to understand host–microbe interactions under warming. The Journal of Experimental Biology 226: jeb245191. https://doi.org/10.1242/jeb.245191
- Frago E, Dicke M, Godfray HCJ (2012) Insect symbionts as hidden players in insect–plant interactions. Trends in Ecology & Evolution 27: 705–711. https://doi.org/10.1016/j.tree.2012.08.013
- Garrido M, Veiga J, Garrigós M, Martínez-de la Puente J (2023) The interplay between vector microbial community and pathogen transmission on the invasive Asian tiger mosquito, Aedes albopictus: Current knowledge and future directions. Frontiers in Microbiology 14: 1208633. https://doi.org/10.3389/fmicb.2023.1208633
- Gerardo NM, Hoang KL, Stoy KS (2020) Evolution of animal immunity in the light of beneficial symbioses. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 375: 20190601. https://doi.org/10.1098/rstb.2019.0601
- Goddard-Dwyer M, López-Legentil S, Erwin PM (2021) Microbiome variability across the native and invasive ranges of the ascidian Clavelina oblonga. Applied and Environmental Microbiology 87: 1–14. https://doi.org/10.1128/AEM.02233-20
- Grbin D, Geček S, Miljanović A, Pavić D, Hudina S, Žučko J, Rieder J, Pisano SRR, Adrian-Kalchhauser I, Bielen A (2023) Comparison of exoskeleton microbial communities of co-occurring native and invasive crayfish species. Journal of Invertebrate Pathology 201: 107996. https://doi.org/10.1016/j.jip.2023.107996
- Gruber M, Quinn O, Baty J, Dobelmann J, Haywood J, Wenseleers T, Lester P (2019) Fitness and microbial networks of the common wasp, Vespula vulgaris (Hymenoptera: Vespidae), in its native and introduced ranges. Ecological Entomology 44: 512–523. https://doi.org/10.1111/een.12732
- Hafsi A, Delatte H (2023) Enterobactereaceae symbiont as facilitators of biological invasion: Review of Tephritidae fruit flies. Biological Invasions 25: 991–1006. https://doi.org/10.1007/s10530-022-02960-x
- Hall L, Nichols C, Martelli F, Leng J, Shuttleworth C, La Ragione R (2024) Significant differences in the caecal bacterial microbiota of red and grey squirrels in Britain. Journal of Medical Microbiology 73: 001793. https://doi.org/10.1099/jmm.0.001793
- Härer A, Rennison DJ (2023) The biogeography of host-associated bacterial microbiomes: Revisiting classic biodiversity patterns. Global Ecology and Biogeography 32: 931–944. https://doi.org/10.1111/geb.13675
- Hassan M, Harmelin-Vivien M, Bonhomme F (2003) Lessepsian invasion without bottleneck: Example of two rabbitfish species (Siganus rivulatus and Siganus luridus). Journal of Experimental Marine Biology and Ecology 291: 219–232. https://doi.org/10.1016/S0022-0981(03)00139-4
- Hernández AM, Alcaraz LD, Hernández-Álvarez C, Romero MF, Jara-Servín A, Barajas H, Ramírez CM, Peimbert M (2024) Revealing the microbiome diversity and biocontrol potential of field Aedes ssp.: Implications for disease vector management. PLoS ONE 19: e0302328. https://doi.org/10.1371/journal.pone.0302328
- Inderjit, van der Putten WH (2010) Impacts of soil microbial communities on exotic plant invasions. Trends in Ecology & Evolution 25: 512–519. https://doi.org/10.1016/j.tree.2010.06.006
- Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, DeSantis TZ (2016) Piphillin: Improved prediction of metagenomic content by direct inference from human microbiomes. PLoS ONE 11: e0166104. https://doi.org/10.1371/journal.pone.0166104
- Jang J, Xiong X, Liu C, Yoo K, Ishii S (2022) Invasive earthworms alter forest soil microbiomes and nitrogen cycling. Soil Biology & Biochemistry 171. https://doi.org/10.1016/j.soilbio.2022.108724
- Jaspers C, Weiland-Bräuer N, Fischer MA, Künzel S, Schmitz RA, Reusch TBH (2019) Microbiota differences of the comb jelly Mnemiopsis leidyi in native and invasive sub-populations. Frontiers in Marine Science 6: 635. https://doi.org/10.3389/fmars.2019.00635
- Jaspers C, Weiland-Bräuer N, Rühlemann MC, Baines JF, Schmitz RA, Reusch TBH (2020) Differences in the microbiota of native and non-indigenous gelatinous zooplankton organisms in a low saline environment. The Science of the Total Environment 734: 139471. https://doi.org/10.1016/j.scitotenv.2020.139471
- Johnson KV-A, Foster KR (2018) Why does the microbiome affect behaviour? Nature Reviews. Microbiology 16: 647–655. https://doi.org/10.1038/s41579-018-0014-3
- Kaur R, Shropshire JD, Cross KL, Leigh B, Mansueto AJ, Stewart V, Bordenstein SR, Bordenstein SR (2021) Living in the endosymbiotic world of Wolbachia: A centennial review. Cell Host & Microbe 29: 879–893. https://doi.org/10.1016/j.chom.2021.03.006
- Kenis M, Benelli G, Biondi A, Calatayud P, Day R, Desneux N, Harrison R, Kriticos D, Rwomushana I, van den Berg J, Verheggen F, Zhang Y, Agboyi L, Ahissou R, Ba M, Bernal J, Bueno A, Carriére Y, Carvalho G, Chen X, Cicero L, du Plessis H, Early R, Fallet P, Fiaboe K, Firake D, Goergen G, Groot A, Guedes R, Gupta A, Hu G, Huang F, Jaber L, Malo E, McCarthy C, Meagher R, Mohamed S, Sanchez D, Nagoshi R, Nègre N, Niassy S, Ota N, Nyamukondiwa C, Omoto C, Palli S, Pavela R, Ramirez-Romero R, Rojas J, Subramanian S, Tabashnik B, Tay W, Virla E, Wang S, Williams T, Zang L, Zhang L, Wu K (2023) Invasiveness, biology, ecology, and management of the fall armyworm, Spodoptera frugiperda. Entomologia Generalis 43: 187–241. https://doi.org/10.1127/entomologia/2022/1659
- Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall L-I, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC (2018) Best practices for analysing microbiomes. Nature Reviews. Microbiology 16: 410–422. https://doi.org/10.1038/s41579-018-0029-9
- Koh A, Bäckhed F (2020) From association to causality: The role of the gut microbiota and its functional products on host metabolism. Molecular Cell 78: 584–596. https://doi.org/10.1016/j.molcel.2020.03.005
- Kohl KD (2017) An introductory “How-to” guide for incorporating microbiome research into integrative and comparative biology. Integrative and Comparative Biology 57: 674–681. https://doi.org/10.1093/icb/icx013
- Kolodny O, Schulenburg H (2020) Microbiome-mediated plasticity directs host evolution along several distinct time scales. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 375: 20190589. https://doi.org/10.1098/rstb.2019.0589
- Koskella B, Bergelson J (2020) The study of host–microbiome (co)evolution across levels of selection. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 375: 20190604. https://doi.org/10.1098/rstb.2019.0604
- Lamer JT, Ruebush BC, Arbieva ZH, McClelland MA, Epifanio JM, Sass GG (2015) Diagnostic SNPs reveal widespread introgressive hybridization between introduced bighead and silver carp in the Mississippi River Basin. Molecular Ecology 24: 3931–3943. https://doi.org/10.1111/mec.13285
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology 31: 814–821. https://doi.org/10.1038/nbt.2676
- Larsen PA, Matocq MD (2019) Emerging genomic applications in mammalian ecology, evolution, and conservation. Journal of Mammalogy 100: 786–801. https://doi.org/10.1093/jmammal/gyy184
- Leonard A, Abalos J, Adhola T, Aguirre W, Aizpurua O, Ali S, Andreone F, Aubret F, Ávila-Palma HD, Alcantara LFB, Beltrán JF, Berg R, Berg TB, Bertolino S, Blumstein DT, giv B, Borowski Z, Boubli JP, Büchner S, Cabido C, Camacho C, Chaparro JC, Charmantier A, D’Elía G, Silva LP da, Dalsgaard B, Franceschi C de, Cruz F de la, Sancha NU de la, Denoël M, Eisenhofer R, Feiner N, Fernandes JM, Figuerola J, Fusani L, Gangoso L, García-Roa R, Gasperini S, Gaun N, Thomas M, Gilbert P, Gomez-Mestre I, Graves GR, Groombridge J, Hardouin EA, Hernández M, M LGH, Hodder K, Hosner PA, Hurtado N, Juste J, Knowles SCL, Kohl KD, Korine C, Kornilev YV, Kramer-Schadt S, Lambin X, Lattenkamp EZ, Lauritsen J, Li G, López CM, Baucells AL, Cohen TM, Manzo E, Marteau M, Martin LB, Bideguren GM, Mazzamuto MV, Monadjem A, Nietlisbach P, Øksnebjerg DB, Packer JG, Pepke ML, Peralta-Sánchez JM, Perdomo A, Lanuza GP i de, Pietroni C, Poyet M, Rahbek C, Ramaraj T, Caldas YR, Razgour O, Rebelo H, Reif J, Rimbach R, Rocha R, Rocha RG, Fernandes CR, Romeo C, Ruuskanen S, Sakaluk SK, Santicchia F, Sarraude T, Sørås R, Spada M, Steele MA, Stothart MR, Sunje E, Sutton AO, Szulkin M, Takahata Y, Thompson CF, Thorup K, Tomazetto G, Torrent L, Toshkova N, Tranquillo C, Turcios-Casco MA, Uller T, Riemsdijk I van, Velo-Antón G, Verbeylen G, Videvall E, Voigt CC, Wauters LA, Wellenreuther M, Yanchukov A, Alberdi A (2025) A global initiative for ecological and evolutionary hologenomics. Trends in Ecology & Evolution 39: 610–620. https://doi.org/10.1016/j.tree.2024.03.005
- Leonhardt F, Keller A, Arranz Aveces C, Ernst R (2023) From alien species to alien communities: Host- and habitat-associated microbiomes in an alien amphibian. Microbial Ecology 86: 2373–2385. https://doi.org/10.1007/s00248-023-02227-5
- Lester P, Bosch P, Gruber M, Kapp E, Peng L, Brenton-Rule E, Buchanan J, Stanislawek W, Archer M, Corley J, Masciocchi M, Van Oystaeyen A, Wenseleers T (2015) No evidence of enemy release in pathogen and microbial communities of common wasps (Vespula vulgaris) in their native and introduced range. PLoS ONE 10: e0121358. https://doi.org/10.1371/journal.pone.0121358
- Lester PJ, Sébastien A, Suarez AV, Barbieri RF, Gruber MAM (2017) Symbiotic bacterial communities in ants are modified by invasion pathway bottlenecks and alter host behavior. Ecology 98: 861–874. https://doi.org/10.1002/ecy.1714
- Levac D, Colquhoun H, O’Brien KK (2010) Scoping studies: Advancing the methodology. Implementation Science 5: 69. https://doi.org/10.1186/1748-5908-5-69
- Levin D, Raab N, Pinto Y, Rothschild D, Zanir G, Godneva A, Mellul N, Futorian D, Gal D, Leviatan S, Zeevi D, Bachelet I, Segal E (2021) Diversity and functional landscapes in the microbiota of animals in the wild. Science 372: eabb5352. https://doi.org/10.1126/science.abb5352
- Liu LJ, Martinez-Sañudo I, Mazzon L, Prabhakar CS, Girolami V, Deng YL, Dai Y, Li ZH (2016) Bacterial communities associated with invasive populations of Bactrocera dorsalis (Diptera: Tephritidae) in China. Bulletin of Entomological Research 106: 718–728. https://doi.org/10.1017/S0007485316000390
- Lockwood JL, Cassey P, Blackburn T (2005) The role of propagule pressure in explaining species invasions. Trends in Ecology & Evolution 20: 223–228. https://doi.org/10.1016/j.tree.2005.02.004
- Lu M, Hulcr J, Sun J (2016) The role of symbiotic microbes in insect invasions. Annual Review of Ecology, Evolution, and Systematics 47: 487–505. https://doi.org/10.1146/annurev-ecolsys-121415-032050
- Mackie RI (2002) Mutualistic fermentative digestion in the gastrointestinal tract: diversity and evolution1. Integrative and Comparative Biology 42: 319–326. https://doi.org/10.1093/icb/42.2.319
- MacLeod CJ, Paterson AM, Tompkins DM, Duncan RP (2010) Parasites lost - do invaders miss the boat or drown on arrival? Ecology Letters 13: 516–527. https://doi.org/10.1111/j.1461-0248.2010.01446.x
- Madhusoodanan J (2019) Do hosts and their microbes evolve as a unit? Proceedings of the National Academy of Sciences of the United States of America 116: 14391–14394. https://doi.org/10.1073/pnas.1908139116
- Malacrinò A, Sadowski VA, Martin TK, de Oliveira NC, Brackett IJ, Feller JD, Harris KJ, Heredia OC, Vescio R, Bennett AE (2020) Biological invasions alter environmental microbiomes: A meta-analysis. PLoS ONE 15: e0240996. https://doi.org/10.1371/journal.pone.0240996
- Maritan E, Quagliariello A, Frago E, Patarnello T, Martino ME (2024) The role of animal hosts in shaping gut microbiome variation. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 379: 20230071. https://doi.org/10.1098/rstb.2023.0071
- Martin Bideguren G, Razgour O, Alberdi A (2024) Quantitative synthesis of microbe-driven acclimation and adaptation in wild vertebrates. Evolutionary Applications 17: e70025. https://doi.org/10.1111/eva.70025
- Martinez-Sañudo I, Simonato M, Squartini A, Mori N, Marri L, Mazzon L (2018) Metagenomic analysis reveals changes of the Drosophila suzukii microbiota in the newly colonized regions. Insect Science 25: 833–846. https://doi.org/10.1111/1744-7917.12458
- McKenzie VJ, Song SJ, Delsuc F, Prest TL, Oliverio AM, Korpita TM, Alexiev A, Amato KR, Metcalf JL, Kowalewski M, Avenant NL, Link A, Di Fiore A, Seguin-Orlando A, Feh C, Orlando L, Mendelson JR, Sanders J, Knight R (2017) The effects of captivity on the mammalian gut microbiome. Integrative and Comparative Biology 57: 690–704. https://doi.org/10.1093/icb/icx090
- McLaren MR, Callahan BJ (2020) Pathogen resistance may be the principal evolutionary advantage provided by the microbiome. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 375: 20190592. https://doi.org/10.1098/rstb.2019.0592
- McLean MA, Migge-Kleian S, Parkinson D (2006) Earthworm invasions of ecosystems devoid of earthworms: Effects on soil microbes. Biological Invasions 8: 1257–1273. https://doi.org/10.1007/s10530-006-9020-x
- Meng Q-Y, Mo D-M, Li H, Wang W-L, Lu H-L (2023) Divergent responses in the gut microbiome and liver metabolome to ammonia stress in three freshwater turtles. The Science of the Total Environment 859. https://doi.org/10.1016/j.scitotenv.2022.160372
- Minard G, Tran FH, Van VT, Goubert C, Bellet C, Lambert G, Kim KLH, Thuy THT, Mavingui P, Valiente Moro C (2015) French invasive Asian tiger mosquito populations harbor reduced bacterial microbiota and genetic diversity compared to Vietnamese autochthonous relatives. Frontiers in Microbiology 6: 970. https://doi.org/10.3389/fmicb.2015.00970
- Moeller AH, Sanders JG (2020) Roles of the gut microbiota in the adaptive evolution of mammalian species. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 375: 20190597. https://doi.org/10.1098/rstb.2019.0597
- Moeller AH, Suzuki TA, Lin D, Lacey EA, Wasser SK, Nachman MW (2017) Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proceedings of the National Academy of Sciences of the United States of America 114: 13768–13773. https://doi.org/10.1073/pnas.1700122114
- Moraïs S, Mizrahi I (2019) The road not taken: The rumen microbiome, functional groups, and community states. Trends in Microbiology 27: 538–549. https://doi.org/10.1016/j.tim.2018.12.011
- Moran NA, Ochman H, Hammer TJ (2019) Evolutionary and ecological consequences of gut microbial communities. Annual Review of Ecology, Evolution, and Systematics 50: 451–475. https://doi.org/10.1146/annurev-ecolsys-110617-062453
- Mowery MA (2024) Endosymbiont diversity across native and invasive brown widow spider populations. Scientific Reports 14: 8556. https://doi.org/10.1038/s41598-024-58723-2
- Neu AT, Allen EE, Roy K (2021) Defining and quantifying the core microbiome: Challenges and prospects. Proceedings of the National Academy of Sciences of the United States of America 118: e2104429118. https://doi.org/10.1073/pnas.2104429118
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S (2012) Host-gut microbiota metabolic interactions. Science 336: 1262–1267. https://doi.org/10.1126/science.1223813
- Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A (2016) Rayyan—A web and mobile app for systematic reviews. Systematic Reviews 5: 210. https://doi.org/10.1186/s13643-016-0384-4
- Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T, Loder EW, Mayo-Wilson E, McDonald S, McGuinness LA, Stewart LA, Thomas J, Tricco AC, Welch VA, Whiting P, Moher D (2021) The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372: n71. https://doi.org/10.1136/bmj.n71
- Paudel S, Longcore T, MacDonald B, McCormick MK, Szlavecz K, Wilson GWT, Loss SR (2016) Belowground interactions with aboveground consequences: Invasive earthworms and arbuscular mycorrhizal fungi. Ecology 97: 605–614. https://doi.org/10.1890/15-1085
- Pérez-Cobas AE, Gomez-Valero L, Buchrieser C (2020) Metagenomic approaches in microbial ecology: An update on whole-genome and marker gene sequencing analyses. Microbial Genomics 6: mgen000409. https://doi.org/10.1099/mgen.0.000409
- Perlman D, Martínez-Álvaro M, Moraïs S, Altshuler I, Hagen LH, Jami E, Roehe R, Pope PB, Mizrahi I (2022) Concepts and consequences of a core gut microbiota for animal growth and development. Annual Review of Animal Biosciences 10: 177–201. https://doi.org/10.1146/annurev-animal-013020-020412
- Peters MDJ, Marnie C, Tricco AC, Pollock D, Munn Z, Alexander L, McInerney P, Godfrey CM, Khalil H (2020) Updated methodological guidance for the conduct of scoping reviews. JBI Evidence Synthesis 18: 2119. https://doi.org/10.11124/JBIES-20-00167
- Pietroni C, Gaun N, Leonard A, Lauritsen J, Martin-Bideguren G, Odriozola I, Aizpurua O, Alberdi A, Eisenhofer R (2024) Hologenomic data generation and analysis in wild vertebrates. Methods in Ecology and Evolution 16: 97–107. https://doi.org/10.1111/2041-210X.14456
- Pyšek P, Hulme PE, Simberloff D, Bacher S, Blackburn TM, Carlton JT, Dawson W, Essl F, Foxcroft LC, Genovesi P, Jeschke JM, Kühn I, Liebhold AM, Mandrak NE, Meyerson LA, Pauchard A, Pergl J, Roy HE, Seebens H, van Kleunen M, Vilà M, Wingfield MJ, Richardson DM (2020) Scientists’ warning on invasive alien species. Biological Reviews of the Cambridge Philosophical Society 95: 1511–1534. https://doi.org/10.1111/brv.12627
- Qin Y, Krosch MN, Schutze MK, Zhang Y, Wang X, Prabhakar CS, Susanto A, Hee AKW, Ekesi S, Badji K, Khan M, Wu J, Wang Q, Yan G, Zhu L, Zhao Z, Liu L, Clarke AR, Li Z (2018) Population structure of a global agricultural invasive pest, Bactrocera dorsalis (Diptera: Tephritidae). Evolutionary Applications 11: 1990–2003. https://doi.org/10.1111/eva.12701
- Qu Y, Wu Y, Zhao Y, Lin L, Du Y, Li P, Li H, Ji X (2020) The invasive red-eared slider turtle is more successful than the native Chinese three-keeled pond turtle: Evidence from the gut microbiota. PeerJ 8: e10271. https://doi.org/10.7717/peerj.10271
- Quince C, Walker AW, Simpson JT, Loman NJ, Segata N (2017) Shotgun metagenomics, from sampling to analysis. Nature Biotechnology 35: 833–844. https://doi.org/10.1038/nbt.3935
- R Core Team (2023) R: A language and environment for statistical computing.
- Reese AT, Dunn RR (2018) Drivers of microbiome biodiversity: A review of general rules, feces, and ignorance. mBio 9: 10. https://doi.org/10.1128/mBio.01294-18
- Risely A (2020) Applying the core microbiome to understand host–microbe systems. Journal of Animal Ecology 89: 1549–1558. https://doi.org/10.1111/1365-2656.13229
- Rosso F, Tagliapietra V, Albanese D, Pindo M, Baldacchino F, Arnoldi D, Donati C, Rizzoli A (2018) Reduced diversity of gut microbiota in two Aedes mosquitoes species in areas of recent invasion. Scientific Reports 8: 16091. https://doi.org/10.1038/s41598-018-34640-z
- Rothman J, Loope K, McFrederick Q, Rankin E (2021) Microbiome of the wasp Vespula pensylvanica in native and invasive populations, and associations with Moku virus. PLoS ONE 16: e0255463. https://doi.org/10.1371/journal.pone.0255463
- Santos B, Bletz MC, Sabino-Pinto J, Cocca W, Solofoniaina Fidy JF, Freeman KLM, Kuenzel S, Ndriantsoa S, Noel J, Rakotonanahary T, Vences M, Crottini A (2021) Characterization of the microbiome of the invasive Asian toad in Madagascar across the expansion range and comparison with a native co-occurring species. PeerJ 9: e11532. https://doi.org/10.7717/peerj.11532
- Saul W-C, Jeschke JM (2015) Eco-evolutionary experience in novel species interactions. Ecology Letters 18: 236–245. https://doi.org/10.1111/ele.12408
- Saul W-C, Jeschke JM, Heger T (2013) The role of eco-evolutionary experience in invasion success. NeoBiota 17: 57–74. https://doi.org/10.3897/neobiota.17.5208
- Seebens H, Bacher S, Blackburn TM, Capinha C, Dawson W, Dullinger S, Genovesi P, Hulme PE, van Kleunen M, Kühn I, Jeschke JM, Lenzner B, Liebhold AM, Pattison Z, Pergl J, Pyšek P, Winter M, Essl F (2021) Projecting the continental accumulation of alien species through to 2050. Global Change Biology 27: 970–982. https://doi.org/10.1111/gcb.15333
- Simberloff D (2006) Invasional meltdown 6 years later: Important phenomenon, unfortunate metaphor, or both? Ecology Letters 9: 912–919. https://doi.org/10.1111/j.1461-0248.2006.00939.x
- Stevens JL, Olson JB (2013) Invasive lionfish harbor a different external bacterial community than native Bahamian fishes. Coral Reefs 32: 1113–1121. https://doi.org/10.1007/s00338-013-1072-7
- Stevens JL, Olson JB (2015) Bacterial communities associated with lionfish in their native and invaded ranges. Marine Ecology Progress Series 531: 253–262. https://doi.org/10.3354/meps11323
- Stevens J, Jackson R, Olson J (2016) Bacteria associated with lionfish (Pterois volitans/miles complex) exhibit antibacterial activity against known fish pathogens. Marine Ecology Progress Series 558: 167–180. https://doi.org/10.3354/meps11789
- Suarez AV, Tsutsui ND (2008) The evolutionary consequences of biological invasions. Molecular Ecology 17: 351–360. https://doi.org/10.1111/j.1365-294X.2007.03456.x
- Suzuki TA, Ley RE (2020) The role of the microbiota in human genetic adaptation. Science. https://doi.org/10.1126/science.aaz6827
- Szklarzewicz T, Świerczewski D, Stroiński A, Michalik A (2020) Conservatism and stability of the symbiotic system of the invasive alien treehopper Stictocephala bisonia (Hemiptera, Cicadomorpha, Membracidae). Ecological Entomology 45: 876–885. https://doi.org/10.1111/een.12861
- Torchin ME, Lafferty KD, Dobson AP, McKenzie VJ, Kuris AM (2003) Introduced species and their missing parasites. Nature 421: 628–630. https://doi.org/10.1038/nature01346
- Traveset A, Richardson DM (2014) Mutualistic interactions and biological invasions. Annual Review of Ecology, Evolution, and Systematics 45: 89–113. https://doi.org/10.1146/annurev-ecolsys-120213-091857
- Tuerlings T, Hettiarachchi A, Joossens M, Geslin B, Vereecken NJ, Michez D, Smagghe G, Vandamme P (2023) Microbiota and pathogens in an invasive bee: Megachile sculpturalis from native and invaded regions. Insect Molecular Biology 32: 544–557. https://doi.org/10.1111/imb.12849
- Utermann C, Blümel M, Busch K, Buedenbender L, Lin Y, Haltli B, Kerr R, Briski E, Hentschel U, Tasdemir D (2020) Comparative microbiome and metabolome analyses of the marine tunicate ciona intestinalis from native and invaded habitats. Microorganisms 8(12): 2022. https://doi.org/10.3390/microorganisms8122022
- Vaelli PM, Theis KR, Williams JE, O’Connell LA, Foster JA, Eisthen HL (2020) The skin microbiome facilitates adaptive tetrodotoxin production in poisonous newts. eLife 9: e53898. https://doi.org/10.7554/eLife.53898
- van Leeuwen PML, Schulte-Hostedde AI, Fournier-Chambrillon C, Fournier P, Pigneur L-M, Aranda CM, Urra-Maya F, Michaux JR (2023) A microbial tale of farming, invasion and conservation: On the gut bacteria of European and American mink in Western Europe. Biological Invasions 25: 1693–1709. https://doi.org/10.1007/s10530-023-03007-5
- Vasconcelos DS, Harris DJ, Damas-Moreira I, Pereira A, Xavier R (2023) Factors shaping the gut microbiome of five species of lizards from different habitats. PeerJ 11: e15146. https://doi.org/10.7717/peerj.15146
- Wagener C, du Plessis M, Measey J (2022) Invasive amphibian gut microbiota and functions shift differentially in an expanding population but remain conserved across established populations. Microbial Ecology 84: 1042–1054. https://doi.org/10.1007/s00248-021-01896-4
- Wang Y, Li Z, Zhao Z (2023) Population mixing mediates the intestinal flora composition and facilitates invasiveness in a globally invasive fruit fly. Microbiome 11: 213. https://doi.org/10.1186/s40168-023-01664-1
- Wilches DM, Laird RA, Fields PG, Coghlin P, Floate KD (2018) Spiroplasma dominates the microbiome of khapra beetle: Comparison with a congener, effects of life stage and temperature. Symbiosis 76: 277–291. https://doi.org/10.1007/s13199-018-0560-5
- Williams CE, Hammer TJ, Williams CL (2024) Diversity alone does not reliably indicate the healthiness of an animal microbiome. The ISME Journal 18: wrae133. https://doi.org/10.1093/ismejo/wrae133
- Zhang Z, Liu Y, Brunel C, van Kleunen M (2020) Soil-microorganism-mediated invasional meltdown in plants. Nature Ecology & Evolution 4: 1612–1621. https://doi.org/10.1038/s41559-020-01311-0
- Zhang L, Yang Z, Yang F, Wang G, Zeng M, Zhang Z, Yang M, Wang Z, Li Z (2023) Gut microbiota of two invasive fishes respond differently to temperature. Frontiers in Microbiology 14: 1087777. https://doi.org/10.3389/fmicb.2023.1087777
- Zhang S, Song F, Wang J, Li X, Zhang Y, Zhou W, Xu L (2024) Gut microbiota facilitate adaptation of invasive moths to new host plants. The ISME Journal 18: wrae031. https://doi.org/10.1093/ismejo/wrae031
- Zhu L, Zhang Z, Chen H, Lamer J, Wang J, Wei W, Fu L, Tang M, Wang C, Lu G (2021) Gut microbiomes of bigheaded carps and hybrids provide insights into invasion: A hologenome perspective. Evolutionary Applications 14: 735–745. https://doi.org/10.1111/eva.13152
- Zhu Y-X, Chang Y-W, Wen T, Yang R, Wang Y-C, Wang X-Y, Lu M-X, Du Y-Z (2022) Species identity dominates over environment in driving bacterial community assembly in wild invasive leaf miners. Microbiology Spectrum 10. https://doi.org/10.1128/spectrum.00266-22
- Zilber-Rosenberg I, Rosenberg E (2008) Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiology Reviews 32: 723–735. https://doi.org/10.1111/j.1574-6976.2008.00123.x
- Zoelzer F, Burger AL, Dierkes PW (2021) Unraveling differences in fecal microbiota stability in mammals: From high variable carnivores and consistently stable herbivores. Animal Microbiome 3: 77. https://doi.org/10.1186/s42523-021-00141-0
- Zuo C, Ma P, Ma X, Zhu Y, Yan S, Zhang Z (2024) Integrated metagenomic and metabolomic analysis on two competing mussels, Mytella strigata and Perna viridis, in China. Animals 14: 918. https://doi.org/10.3390/ani14060918
Supplementary materials
PRISMA flowchart
Data type: pdf
Explanation note: PRISMA flowchart illustrating the systematic review process.
List of reviewed articles
Data type: csv
Explanation note: Full list of the 147 articles included in the review, their metadata and associated descriptors.
Study design descriptors
Data type: pdf
Explanation note: Description of the categories used to define the study design of the assessed articles.